
NanoCommons Knowledge Base and Data Warehouse powered by 
NanoCommons is providing a common user interface for accessing data from different existing resources and is also building a home for data, which is not publicly released otherwise.
Table of contents
Access
NanoCommons knowledge base and data warehouse
Short manual (from NanoCommons deliverable report, June 2022)
Online manual (login to NanoCommons KB required)
The NanoCommons data infrastructure was designed to become the one-stop information source on nanosafety. It is providing access according to the FAIR principles by:
- Integrating data directly into the NanoCommons data warehouse, which was not publicly available until then and might come from projects or individual users.
- Cataloguing and making available existing databases and data warehouses from European and international projects specialised for specific types of data
To be able to distinguish between these two tasks, the platform is referred to by the two terms:
- NanoCommons Knowledge Base (KB) establishes the links to the data sources including the NanoCommons Data Warehouse and provides the user interface and application programming interfaces (APIs) for advanced data visualisation, searching, browsing and accessing features.
- NanoCommons Data Warehouse (DW) is a part of the NanoCommons KB, containing the data that is made publicly available for the first time (and not just linked to) by NanoCommons.
For the data user, the separation between these two parts is not relevant since all data can be search and browsed via the same interface even if there might then be differences in the access. In contrast, data providers will follow different workflows to prepare and upload the data depending on the solution they want to use for storing their data. More information on these workflows is provided in the NanoCommons data workflows section of this handbook.
Integration of project data directly or by linking to other resources
As described above, the NanoCommons KB has two ways, how (meta)data is integrated to then be shown to the users using the common interface. These will be described in more detail using examples from specific EU projects.
Providing data for the first time
The first version of the approach that has evolved to become the NanoCommons KB was established within the Framework Programme 7 (FP7) project NanoMILE which initially included Biomax as a partner to handle the omics datasets, but which quickly identified a significant gap in how NMs data is captured, and in particular identified a gap in terms of the evolution of NMs in contact with biological and environmental milieu, and the wide range of transformation reactions that NMs undergo. Thus, in NanoMILE, the goal was to develop a system that allowed the characteristics of NMs as received to be linked to the properties of the NMs dispersed in different media, and to the characteristics of the NMs following various ageing and transformation processes, such that we could query the data to identify whether, for example, pristine of transformed properties were more predictive of toxicological impact, and whether transformation increased or decreased the “similarity” of NMs of the same NM produced by different routes or with different initial capping agents, for example, and indeed whether NMs of different compositions aged to similar surface compositions and whether this lead to similar fates and toxicities.
A large volume of data was generated in NanoMILE, spanning NMs characterisation, high content screening analysis in human cells and zebrafish embryos, and toxicity and ecotoxicity studies on a small subset of the total NanoMILE NMs library, which consisted of >100 well characterised NMs. However, the processes for integration of the data were not yet developed while NanoMILE was generating its datasets, and thus while some of the data is integrated into the KB, work is underway to integrate the rest of the data. The physico-chemical characterisation data and the high throughput screening data are integrated (and are also published as in Joossens et al., 2019, and the data deposited in the JRC data repository), as are the omics datasets including some protein corona datasets, and the computational data generated from the Quantitative Structure Activity Relationships (QSAR) models.
The Horizon 2020 project NanoFASE built upon the initial NanoMILE database, with the NanoFASE data warehouse being version 2.0 of what is now the NanoCommons KB. NanoFASE focussed entirely on characterising (and developing a model to predict) the transformations of NMs in various environmental compartments, including air, water, sediment, soil, during wastewater treatment / in sludge, during incineration and following uptake into organisms and plants. In addition to the individual compartment / species studies, NanoFASE undertook some ambitious mesocosm studies. As a demonstration study for NanoCommons, detailed data capture templates were developed to support the integration of the NanoFASE mesocosm data into the NanoFASE/NanoCommons database. NanoFASE also focussed on the development of functional assays that can predict key transformations of NMs without needing to run full mesocosms or pilot waste treatment plant scale experiments. The datasets are based on the NIKC template that was developed by NanoCommons partners the Center for the Environmental Implications of Nanotechnology. They are Excel based to facilitate data input by experimentalists and enable quick and easy upload. All templates are ontologically mapped to NanoCommons-incorporated ontologies to facilitate the semantic mapping and subsequent data mining, data harvesting and data re-use.
Integrating and mapping of (meta)data from external data resources
The NanoCommons KB provides a REST Webservice Application Programming Interface (API) and a semantic mapping mechanism (Losko and Heumann, 2017) that in combination allow to connect to existing databases and provide information on available content, generating a searchable catalogue of existing information while actual data access remains with the connected database.
As proof-of-concept, a connection with the AMBIT data warehouse software has been established which is used to create databases for several research projects. Via the NanoCommons KB currently the following are searchable: eNanoMapper, NANoREG, NANoREG2
Extended data upload mechanism and templates
Based on feedback from users of the NanoCommons KB, the standard data upload mechanism and associated templates were extended. The upload now allows batch import of NMs
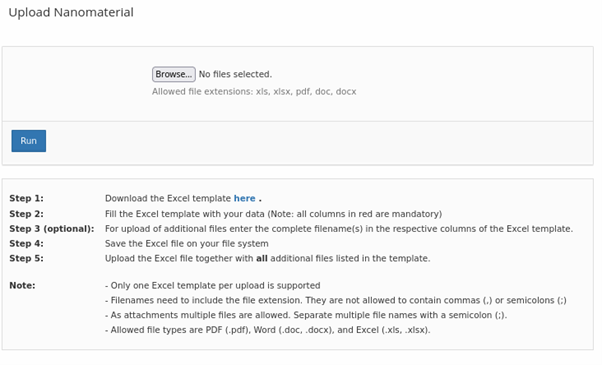
of measurement parameters (“endpoints”)
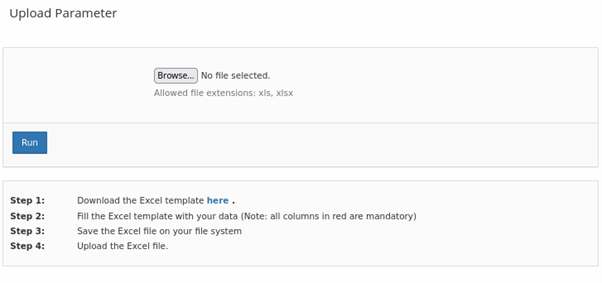
and of actual measurement data
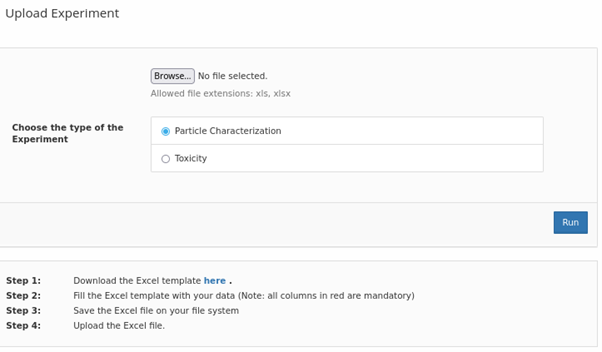
Due to the fast technological and scientific development in this field, the full set of measurement parameters, which have to be accommodated by the NanoCommons infrastructure, cannot be well defined currently and will probably have to continuously be extended in the future. At the same time the infrastructure capability needs to extend beyond simple data archives to encompass also semantically undefined measurement parameters. Therefore an upload and mapping mechanism has been developed, which allows users to dynamically extend the measurement parameters accommodated by the system while at the same time ensuring semantic mapping of new parameters via ontologies.
Training
The session from the 2021 Nanosafety Training School give a motivation on why and information on how to use the Knowledge Base using an in vitro experimental to in silico modelling workflow. Background on the present FAIR data initiative in the context of nanosafety assessment and risk governance for nanomaterials and nanotechnology-based research & development are given first. Then. it covers the data life cycle, explains the concept of metadata completeness and what a data shepherd does or is responsible for. A practical example is worked through investigating the protein corona formation around nanoparticles by a suite of experimental analyses combined with in silico modelling of the protein corona and making use of FAIR data provision using the NanoCommons Knowledge Base. Herein you can define your self-produced nanomaterial and deposit all relevant physicochemical parameters that you have determined. You have access to the ontology terms and can look them up. You also find the NanoXtract TEM image analysis tool as well as the protein corona prediction tool and can start them from within the Knowledge Base.
NanoCommons – Knowledge Base Data Workshop Workshop giving a introduction to the NanoCommons Knowledge Base, presenting applications to mesocosm and in vitro-in silico workflows and describing some additional developments.
Using the NanoCommons Knowledge Base Short introduction to the functionality of the Knowledge Base (at the Jaqpot Hackathon). Much shorter as the above but also a little outdated.
Using the NanoCommons Knowledge portal for templates and annotation upload Support for data preparation and integration into the semantic model of the Knowledge Base.
Descriptions of concepts and implementation
First version of data warehouse and collaborative knowledge infrastructure
Initial Knowledge Infrastructure Functionalities and Services Implemented
References
- Joossens et al., 2019: Joossens, E.; Macko, P.; Palosaari, T.; Gerloff, K.; Ojea-Jiménez, I.; Gilliland, D.; et al. A high throughput imaging database of toxicological effects of nanomaterials tested on HepaRG cells. Sci Data 2019, 6, 46. https://doi.org/10.1038/s41597-019-0053-2.
- Losko and Heumann, 2017: Losko, S.; Heumann, K. Semantic data integration and knowledge management to represent biological network associations. Methods Mol Biol. 2017, 1613, 403–423. https://doi.org/10.1007/978-1-4939-7027-8_16.