
NanoCommons Knowledge Base and Data Warehouse explained
How to get started with the NanoCommons KB user interface.
Knowledge Base user interface
In order to give an overview about how the NanoCommons Knowledge Base (NanoCommons KB) user interface works, we provide in this section a short, screenshot based guide to the information available and the navigation within the user interface. More details on the the BioXMTM software can be found in Losko and Heumann, 2009, while the method for generating the semantic mappings is described in Maier et al., 2011.
Login
Users access the login page to enter the secure, authentication and authorisation based NanoCommons KB (Figure 1). This approach allows the NanoCommons KB to be fully compliant with the General Data Protection Regulations (GDPR), and to provide tailored solutions and support for individual users. Currently the usage is restricted to NanoCommons partners. As a next step, an auto-registration will enable external users to automatically register and access the information and tools as soon as they are approved by NanoCommon partners for public access. Later stages may also implement single sign on with SAML or OAuth2 services.

Figure 1. Login for the NanoCommons Knowledge Base user interface
Home page
You will now be in the Home page (Figure 2). On the left you can find a frame providing navigation to the different types of information integrated into the NanoCommons KB.
On the home page screen is a diagram providing a Tiles based overview of the different Information, Actions and Tools currently available. This interface is highly flexible and will change as the project evolves and new information, functions and tools become available and are offered as Transnational Access (TA) services.
The navigation menu (on the left) provides links to content and resources available in the KB as well as the search and management functionalities. The following sections are available:
- Home — links to the most relevant content in the KB (explained below)
- News — links to the NanoCommons project news feed
- My Profile — interface for managing your personal contact data and login information
- Help — link to the help page providing Frequently Asked Questions (FAQ) and guidance for Users, as well as pointing users to the Help Desk (see below)
- Help Desk — link to the electronic issue tracking
- Data Access — links to the available data sets, with search and report functionalities
- Data Upload — interface for uploading data sets into the KB as well as instructions and guidance for users on how to ontologically annotate their datasets for upload into the KB
- Analysis — links to the available analysis pipelines with information on required data formats
- Browse ontologies — links to the available ontologies with browse and search functionalities, again lined to the guidance for users on how to ontologically annotate their datasets for upload into the KB.
Additional menu items will become available as corresponding material is developed, e.g.:
- Training Materials - links to the training materials, video tutorials and other user support offerings developed by NanoCommons partners or tool providers
- Demonstration Case studies - links to the project, industry and regulatory case studies underway or completed and their outcomes.
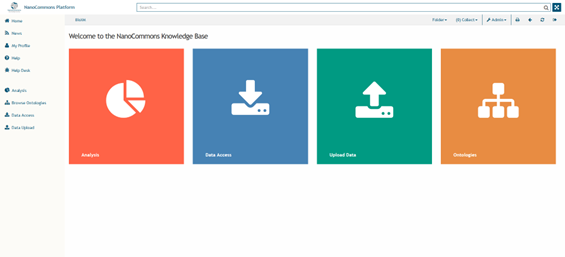
Figure 2. Homepage of the NanoCommons Knowledge Base.
Browsing NMs in the knowledge base
Users can browse through the following information related to nanomaterials (NMs) included in the NanoCommons KB:
- NMs General Data (including supplier information, general description, composition information, and European Registry of Materials (ERM) ID number)
- NMs Characterisation Data (including ageing and transformations)
- NMs Omics Data
- NMs toxicity / ecotoxicity data
- NMs release, exposure and environmental fate data
- NMs Computational nanodescriptors
- To browse the NMs, click the appropriate link. A table of (currently) 416 NMs will be displayed as the result (Figure 3). The table lists the NMs with their respective general information:
- ID (and soon also European Registry ID, a unique and persistent identifier for each NM, including transformed or aged variants, and computational NMs) Chemical Elements including any coatings or capping agents
- Basic characterisation data (e.g. size and shape, crystal phase where relevant)
- Samples (batches or lots) including synthesis date (if available) and/or opening date of bottle (for commercial samples) as part of the Provenance information
- Aliquots (where a larger sample was sub-sampled for distribution to project partners)
- Project-specific name where relevant
- Designator (e.g. Producer’s identifier or code)
- All names (any synonyms or other short codes used by researchers for this NM)
- Description of physical parameters such as size or form
- Aging reaction (if relevant)
- Storage conditions for the samples to minimise ageing during storage
Clicking on a NM ID will display the “Nanomaterial properties report” for that NM. This report contains the available physicochemical and computational characterisation parameters for each material. These will also be linked to the protocols and methods used for the experimental characterisation or computational models used to calculate the theoretical descriptors that will be included into the NanoCommons protocols repository.
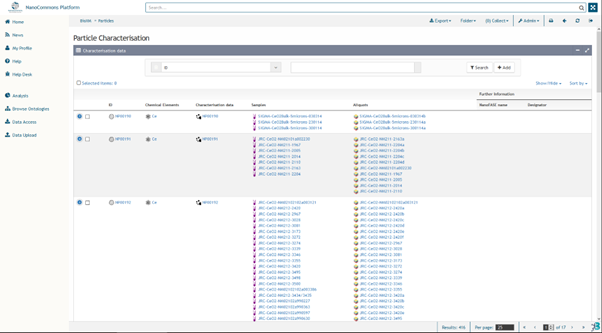
Figure 3. List of NMs currently available in the NanoCommons Knowledge Base, which have come mainly from the NanoMILE and NanoFASE projects to date. A list of literature curated NMs is currently being added, as part of the European Registry of Materials (ERM), developed as part of NanoCommons, and which is currently assigning a persistent and unique Identifier (PId) to each individual NM, including individual batches, aged variants and computationally derived NMs.
Browsing physico-chemical characterization results
To browse NanoCommons Characterization experiments click the “Characterization” link on the “Home” page. The “Particle Characterisation” page, in which the experiments are listed in a table, will be displayed with the characterization data for each NMs (Figure 4).
Typically, information provided includes:
- Size and size distribution (also called polydispersity index, PDI), indicating by which method data was generated and for electron microscopy, the number of NMs that were analysed.
- Shape
- Crystallinity
- Coating composition and how the coating is attached to the NM
- Form in which the NM was supplied (e.g. powder, aqueous dispersion etc.)
- Dispersion liquid
- Redox state (where relevant)
- Zeta potential as a measure of surface charge, which should be accompanied by the pH at which it was measured, and the ionic strength of the measurement liquid.
- Electromobility, which is derived from the zeta potential value
- TEM or STEM derived size information. If available also images of the NM could be stored and made available to calculate an additional set of computational descriptors, utilising two of the tools integrated into NanoCommons, namely NanoXtract and NanoImage, as described in the Nanoinformatics section.
- Energy Band Gap, which is calculated from the UV-Vis spectrum.
However, the availability of these and additional attributes depends on the individual data set, as not all physico-chemical endpoints are relevant to all NMs. For example, redox state is relevant for some metals (e.g. Fe, Ce) but is not relevant for others (e.g. Ti, Si) while some elements can exist in multiple crystal phases (e.g. Ti can exist as anatase, rutile, brookite and amorphous forms) while other elements have only one lattice structure.
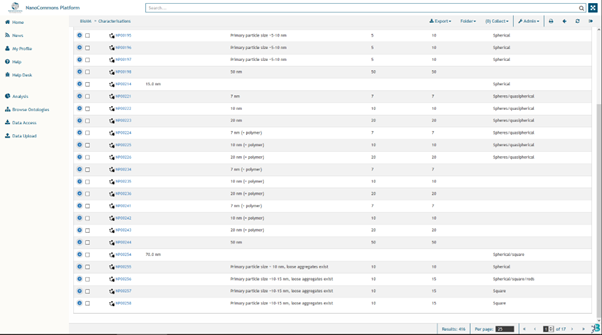
Figure 4. Physico-chemical characterisation available in the NanoCommons KB
Browsing Omics results
To browse NanoCommons Omics experiments click the “Omics” link on the “Home” page. The “RNA-Seq” page in which the experimental results are listed in a table, will be displayed (Figure 5).
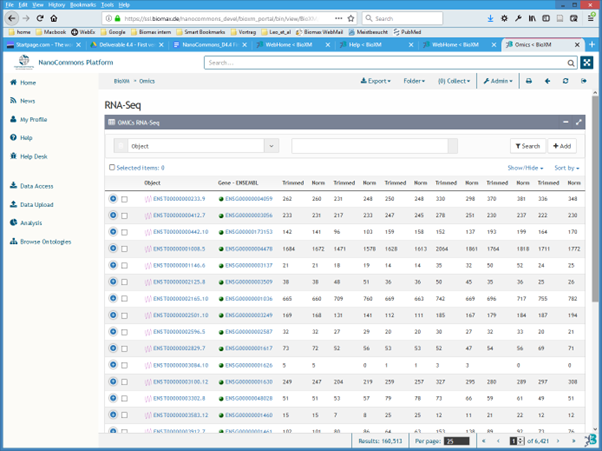
Figure 5. RNA-seq measurements of specific exposure experiments available in the NanoCommons KB
Ontology search tools
To facilitate dataset owners, who are generally experimentalists with limited experience of ontologies or using programming repositories such as GitHub, a simple, user-friendly interface has been implemented that allows users to quickly and easily search for ontological terms and identifiers that match the terms used in their datasets. By annotating the identified ontology terms to their experimental terms, the database then knows exactly where to add each term, and the associated data, into the database to make it easily searchable and retrievable, and to enable combining disparate datasets. Figure 6 shows a screen-shot of the ontology search tool implemented into the NanoCommons KB. A range of relevant ontologies have been incorporated already and, as part of NanoCommons, new terms are being added where needed. More information on the recommended ontologies, the ontology development and the search tools is available in the Ontology section.
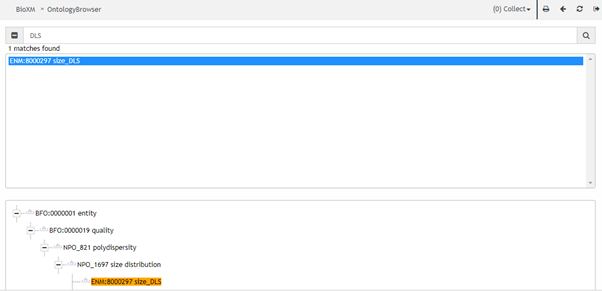
Figure 6: Screenshot of the user-friendly ontology look-up service (BioXM ontology search tool) integrated into the NanoCommons KnowledgeBase (data management module), and integrating the eNanoMapper, the European Materials Modelling Ontology (EMMO), PATO and many other relevant ontologies for NMs, their characterisation, exposure and hazard characterisation and risk assessment.
References
- Losko and Heumann, 2009: Losko, S.; Heumann, K. Semantic data integration and knowledge management to represent biological network associations. Methods Mol Biol. 2009, 563, 241–258. https://doi.org/10.1007/978-1-60761-175-2_13.
- Maier et al., 2011: Maier, D.; Kalus, W.; Wolff. M.; Kalko, S.G.; Roca, J.; Marin de Mas, I.; et al. Knowledge management for systems biology a general and visually driven framework applied to translational medicine. BMC Syst Biol. 2011 5: 38. https://doi.org/10.1186/1752-0509-5-38.