Read-across methodologies
Read-across non-testing strategies are employed for the prediction of NM toxicity, in cases where sufficiently large datasets are not available for the development of reliable nanoQSAR models. This approach is grounded on the empirical knowledge that similar materials may exhibit comparable properties and thus, the estimation of the hazardous effects of non-tested NMs can be achieved using data within a group of comparable NMs.
The European Chemicals Agency (ECHA) through the Read-Across Assessment Framework (RAAF) has introduced a read-across strategy that consists of seven well defined steps (ECHA, 2017), as follows:
- Determination of the structural characteristics of NMs (composition, including surface chemistry and any impurities, size, shape etc.).
- Development of an initial grouping hypothesis that correlates an endpoint (e.g. a toxicity index), to different behavior and reactivity properties, including solubility, zeta potential, dispersibility, hydrophobicity, dustiness, biological activity (redox formation, gene expression etc.), photoreactivity etc. Assignment of the samples to groups.
- Gathering of the above properties (depending on the grouping hypothesis) for each NM.
- Construction of a data matrix including properties and endpoints.
- Assessment of the applicability of the approach using computational techniques (e.g. Principal Components Analysis (PCA), hierarchical clustering, random forests, linear discriminant analysis (LDA) etc.), and data gaps filling. If no regular pattern emerges, an alternative grouping hypothesis must be proposed (step 2).
- In case that the grouping hypothesis is robust, but adequate data are not available, additional testing should be considered to complete the datasets.
- Justification of the method.
A more detailed presentation of RAAF is provided in Figure 1.
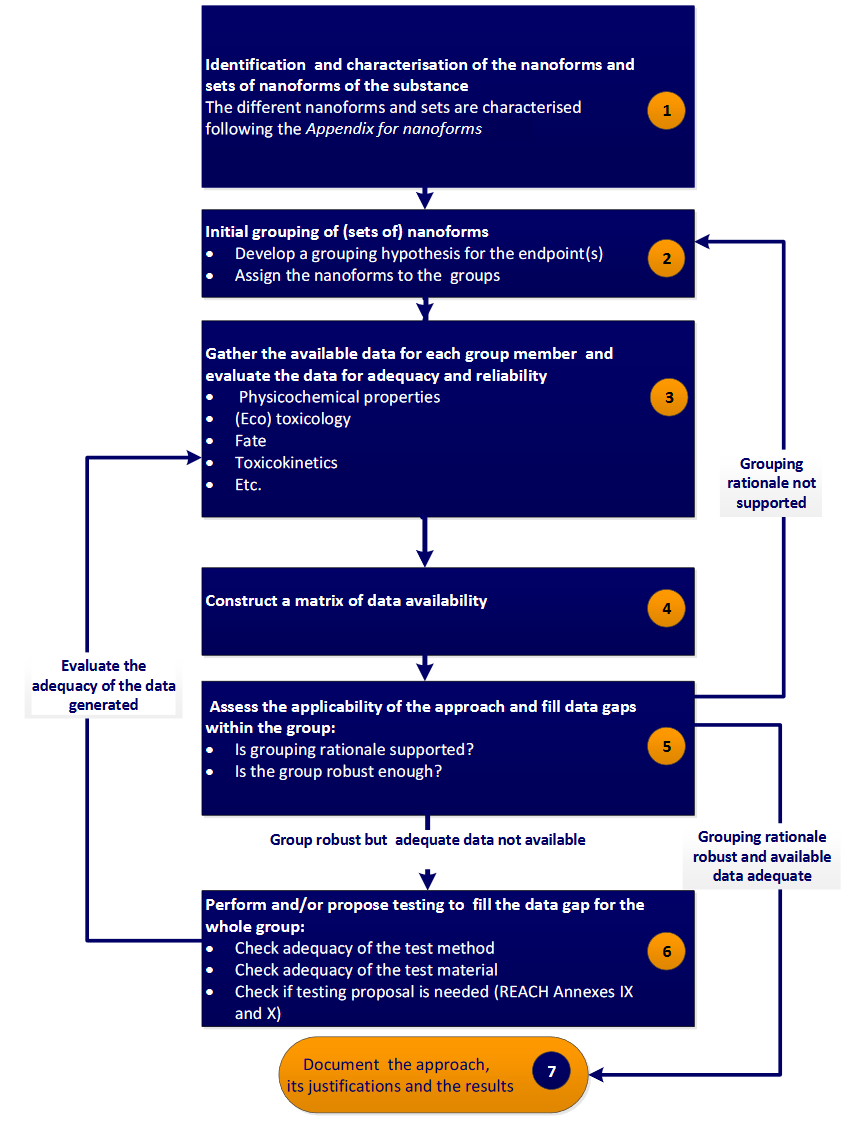
Figure 1. Step-wise grouping/read-across approach proposed by ECHA in Appendix R.6-1 for nanoforms, from the Guidance on QSARs and Grouping of Chemicals (ECHA, 2017)
Grouping/read-across services
NTUA’s toxFlow and Apellis tools
NTUA has designed novel grouping/read-across approaches which automate and optimise the Read-across scheme that has been proposed by ECHA. These approaches have been implemented in the form of web services that have been integrated into the NanoCommons infrastructure:
The toxFlow tool (https://toxflow.jaqpot.org/) is a web based R-Shiny implementation of the abovementioned grouping/read-across approach, where the user can manually select most of the parameters involved in the construction of the read-across problem. More specifically, the toxicity endpoint prediction of the target ENM can be performed using the weighted average of the corresponding values of the neighbor ENMs, i.e. neighboring substances of every target ENM are selected by calculating pairwise similarity measures with all available ENMs and by excluding those ENMs for which the similarity measure does not fulfill a predefined threshold. The proposed workflow assumes that a complete training data set is available, i.e., a set of ENMs for which the toxicity endpoint values are known, as described in Varsou et al., 2017.
The Apellis tool (https://apellis.jaqpot.org/) automates the process of searching over the solution space in order to find the read-across hypothesis that produces the best possible results in terms of prediction accuracy and number of ENMs for which predictions are obtained (Varsou et al., 2019). Thus, it overcomes a main drawback of existing approaches, which are based on manually trying different read-across hypotheses in an iterative, inefficient and time-consuming trial and error fashion. The main outcomes of the method are a reduced set of significant descriptors and a single or multiple threshold value(s) which rigorously define the boundaries around a query ENM, where neighboring ENMs are located. These two different goals are achieved simultaneously through the development of a Mixed Integer NonLinear Programming (MINLP) problem, where the objective is to minimize the Mean Squared Error (MSE) between the experimental values and the produced predictions with respect to selecting the most informative descriptors and defining the neighbor boundaries.
References
- ECHA, 2017: ECHA, Read Across Assessment Framework, Appendix R.6-1 for nanomaterials applicable to the Guidance on QSARs and Grouping of Chemicals. Guidance on information requirements and chemical safety assessment, 2017. https://echa.europa.eu/documents/10162/23036412/appendix_r6_nanomaterials_en.pdf/71ad76f0-ab4c-fb04-acba-074cf045eaaa
- Varsou et al., 2017: Varsou, D.D.; Tsiliki, G.; Nymark, P.; Kohonen, P.; Grafström, R.; Sarimveis, H. toxFlow: a web-based application for read-across toxicity prediction using omics and physicochemical data. Journal of chemical information and modeling 2017, 58(3), 543-549. https://doi.org/10.1021/acs.jcim.7b00160
- Varsou et al., 2019: Varsou, D.D.; Afantitis, A.; Melagraki, G.; & Sarimveis, H. Read-across predictions of nanoparticle hazard endpoints: a mathematical optimization approach. Nanoscale Advances 2019, 1(9), 3485-3498. https://pubs.rsc.org/en/content/articlehtml/2019/na/c9na00242a