NanoCommons Demonstration Case - Alignment of in silico protein corona modelling with experimental analysis
Bio-nano interaction data (characterization of nanomaterials with and without allergens, structural alterations upon nanoparticle binding, selective binding of allergens from protein mixes, investigations of protein orientations when bound to NMs, etc.) are generated in different past and ongoing projects. This demonstration case was designed around this research to see how corona modelling can help to understand the effects. This case has been finalised.
Table of contents
Background and aims
The interaction of allergens with engineered nanoparticles (ENP) and immune effects thereof may have an impact on medical applications and their safety aspects. These are relevant in regard to allergen-specific immunotherapy (AIT) but also for safety assessment at occupational settings where allergens and nanoparticles may come in contact. At PLUS, bio-nano interaction data (characterization of NM with and without allergens, structural alterations upon nanoparticle binding, selective binding of allergens from protein mixes, investigations of protein orientations when bound to NMs, etc.) are generated in different past and ongoing projects. Furthermore, biological effects of allergen-NP conjugates are investigated involving a panel of different in vitro systems.
This demonstration case was designed around this research to see how corona modelling can help to understand the effects. The following aims were defined:
- Aim 1 was to align in silico approaches for protein corona formation with experimental data as this seems to be essential to further optimize future prediction capacities for hazard assessment. Results from in silico protein corona modelling strategies as established at partner UCD of the NanoCommons consortium need to be correlated with the experimental results from the lab at PLUS. The primary focus of this demonstration case was to elucidate a potential selectivity in protein binding, and to investigate whether certain orientations are preferred and whether structural alterations occur upon non-covalent binding to NPs. In case of using allergens as model systems, such conformational alterations might have an impact on the allergenicity of the allergen-NP conjugate due to epitope modification or masking.
- Aim 2 was to harmonise the generated data with and integrate into the NanoCommons Knowledge Base (KB), which was facilitated by partner Biomax.
Key questions
- Can experimentalists apply current in silico protein corona modelling procedures upon knowledge transfer by a short-term TA service?
- Can experimental data on protein corona formation be aligned with current data of current in silico modelling procedures?
- What do we need to further optimize current in silico modelling procedures?
- How can we further optimize knowledge transfer by a short-term TA service or improved training material?
- How can a new type of data, such as the one of structural alterations upon protein corona formation, be uploaded onto the NanoCommons KB?
Results
The primary focus was to investigate potential differences in protein binding to the differently surface-functionalized/coated SiO2 NPs. Therefore, in vitro binding studies, using SDS-PAGE, were performed with an artificial protein mix and a crude birch pollen extract (BPE) interacting with either pristine or APTES-coated SiO2 NPs. To examine the impact of varying environmental conditions on protein corona formation, the couplings were performed in two different environments, pure water and pH 4 citrate buffer. To allow potential alignment of in silico and in vitro results, the ζ-potential values required for in silico prediction modelling with the UnitedAtoms tool (Power at al., 2019) were determined in water and in pH 4 buffer. The calculated binding energies of the modelling approach and the binding capacities according to the in vitro binding studies are listed in Table 1 and 2 to rank the proteins according to their binding affinities. This should approximately also represent the composition of the protein corona.
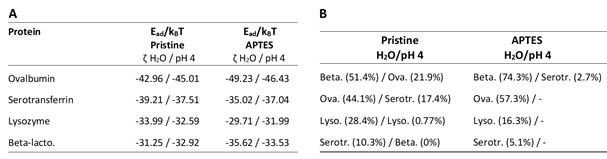 Table 1. Ranking of artificial mix proteins on pristine and APTES-coated SiO2 NPs. A) Minimum adsorption energies (Ead) for proteins of interest on SiO2 NPs. All values are reported in units of kBT (Boltzmann constant at Temperature in Kelvin). ζ-potentials of particles used for modelling were determined in H2O and pH 4 buffer. Proteins are ranked by descending adsorption energy on pristine SiO2 NPs. B) Ranking of proteins according to in vitro binding capacity on SiO2 NPs coupled in H2O or pH 4 buffer. Protein bound to NPs in % of control in brackets. Beta. (Beta-lactoglobulin a, PDB-ID 1CJ5), Ova. (Ovalbumin, PDB-ID 1UHG), Lyso. (Lysozyme, PDB-ID 1DPX), Serotr. (Serotransferrin, PDB-ID 6JAS).
Table 1. Ranking of artificial mix proteins on pristine and APTES-coated SiO2 NPs. A) Minimum adsorption energies (Ead) for proteins of interest on SiO2 NPs. All values are reported in units of kBT (Boltzmann constant at Temperature in Kelvin). ζ-potentials of particles used for modelling were determined in H2O and pH 4 buffer. Proteins are ranked by descending adsorption energy on pristine SiO2 NPs. B) Ranking of proteins according to in vitro binding capacity on SiO2 NPs coupled in H2O or pH 4 buffer. Protein bound to NPs in % of control in brackets. Beta. (Beta-lactoglobulin a, PDB-ID 1CJ5), Ova. (Ovalbumin, PDB-ID 1UHG), Lyso. (Lysozyme, PDB-ID 1DPX), Serotr. (Serotransferrin, PDB-ID 6JAS).
 Table 2. Ranking of BPE proteins on pristine and APTES-coated SiO2 NPs. A) Minimum adsorption energies (Ead) for proteins of interest on SiO2 NPs. All values are reported in units of kBT (Boltzmann constant at Temperature in Kelvin). ζ-potentials of particles used for modelling were determined in H2O and pH 4 buffer. Proteins are ranked by descending adsorption energy on pristine SiO2 NPs. B) Ranking of BPE proteins according to in vitro binding capacity on SiO2 NPs coupled in H2O or pH 4 buffer. Five most prominent bands of BPE gel were chosen for quantitative comparison and according to their molecular weight assigned to the main birch pollen allergens. Protein bound to NPs in % of control in brackets. BPE (birch pollen extract).
Table 2. Ranking of BPE proteins on pristine and APTES-coated SiO2 NPs. A) Minimum adsorption energies (Ead) for proteins of interest on SiO2 NPs. All values are reported in units of kBT (Boltzmann constant at Temperature in Kelvin). ζ-potentials of particles used for modelling were determined in H2O and pH 4 buffer. Proteins are ranked by descending adsorption energy on pristine SiO2 NPs. B) Ranking of BPE proteins according to in vitro binding capacity on SiO2 NPs coupled in H2O or pH 4 buffer. Five most prominent bands of BPE gel were chosen for quantitative comparison and according to their molecular weight assigned to the main birch pollen allergens. Protein bound to NPs in % of control in brackets. BPE (birch pollen extract).
Comparison of the entries of the respective columns A and B of Table 1 and 2 revealed substantial deviations between the experimentally obtained in vitro results and the calculated adsorption affinities of the in silico modelling. The actual composition of the protein corona did not coincide with the predicted composition. Whereas the in vitro results showed clear differences in the proteins´ binding behaviour depending on the environmental pH value, which influences the surface charge of proteins and NPs, and therefore their electrostatic interactions, the in silico results suggested very similar binding affinities of all tested proteins in a quite narrow range regardless of the environmental conditions.
Further in silico modelling approaches were conducted with UnitedAtoms to model the adsorption energies and preferred orientations of the major birch pollen allergen Bet v 1 on the four differently coated SiO2 NPs. Figure 1 shows the most likely orientation of Bet v 1 on (A) pristine (B) APTES- (C) TSUA- and (D) PTMO-coated SiO2. These heatmaps display the predicted adsorption energies for all possible orientations of Bet v 1 on the respective NP surface and Figure 1E lists the minimum adsorption energies of Bet v 1 on the respective NPs with the corresponding angles. The strongest binding of Bet v 1, using ζ-potential values of the NPs determined in H2O, was predicted for APTES-coated SiO2 NPs, whereas the strongest binding was predicted for PTMO-coated NPs, using ζ-potential values determined in pH 4 buffer.
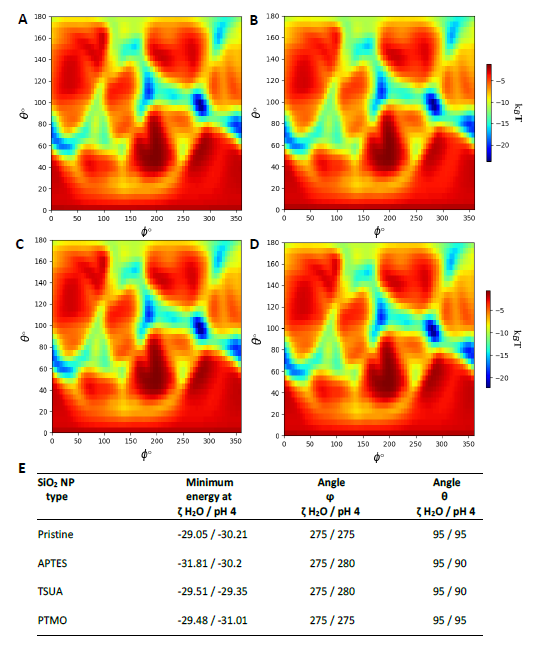 Figure 1. Orientation-specific binding energy of Bet v 1 (PDB-ID 4A88) on SiO2 NPs. (A) pristine (B) APTES- (C) TSUA- (D) PTMO-coated SiO2. ζ-potentials of particles used for modelling were determined in H2O. Adsorption energy reported in units of kBT (Boltzmann constant at Temperature in Kelvin). (E) Minimum adsorption energy and corresponding angles of Bet v 1 interacting with differently coated silica NPs. ζ-potentials of particles used for modelling were determined in H2O and pH 4 buffer. Angles ϕ and θ correspond to specific orientation of the protein on the NP surface, determined by rotation around the Y and Z axes of the corresponding PDB 3D structure of the protein.
Figure 1. Orientation-specific binding energy of Bet v 1 (PDB-ID 4A88) on SiO2 NPs. (A) pristine (B) APTES- (C) TSUA- (D) PTMO-coated SiO2. ζ-potentials of particles used for modelling were determined in H2O. Adsorption energy reported in units of kBT (Boltzmann constant at Temperature in Kelvin). (E) Minimum adsorption energy and corresponding angles of Bet v 1 interacting with differently coated silica NPs. ζ-potentials of particles used for modelling were determined in H2O and pH 4 buffer. Angles ϕ and θ correspond to specific orientation of the protein on the NP surface, determined by rotation around the Y and Z axes of the corresponding PDB 3D structure of the protein.
When investigating the binding behaviour of Bet v 1 and the four differently coated SiO2 NPs in vitro, using BCA-protein assay and SDS-PAGE, the results showed that there were clear differences in the binding capacities of the different NPs (Figure 2). The highest binding capacity was observed in pH 4 citrate buffer, followed by binding in pH 7.4 and pH 9 buffers for all four SiO2 NP types. This coincides with previous findings, showing that the pH optimum for the coupling buffer is determined by the pI value of the respective protein and that the binding is most efficient at a pH value slightly below the pI value of the protein (Lahiri et al., 1999). The TSUA-coated SiO2 NPs, possessing the most negatively charged surface potential of all four investigated NPs in addition to a highly hydrophobic character, showed the highest binding capacity for Bet v 1 in all three examined buffers. This indicates, in accordance with previous findings (Cedervall et al., 2007)(Deng et al., 2013), that electrostatic as well as hydrophobic interactions play a crucial role in the formation of the protein corona.
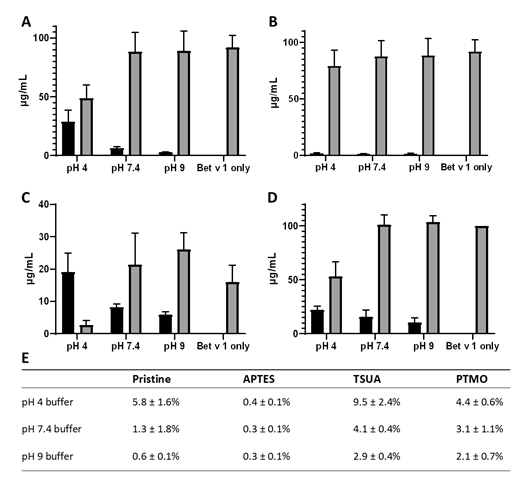 Figure 2. Binding of Bet v 1 on four differently coated SiO2 NPs. (A) pristine (B) APTES- (C) TSUA- (D) PTMO-coated SiO2 NPs. Black bars: bound protein, Grey bars: unbound protein. Coupled in pH 4 citrate buffer, 10 mM; pH 7.4 sodium phosphate buffer, 10 mM; pH 9 Tris buffer, 10 mM. Results of BCA protein assay expressed as µg mL-1 of protein in pellet (bound) or supernatant (unbound); Bet v 1 only as control; Graphs show mean ± SD of three experimental repeats. (E) Binding capacity in weight percent of four different SiO2 NPs with Bet v 1. Mean ± SD of three experimental repeats.
Figure 2. Binding of Bet v 1 on four differently coated SiO2 NPs. (A) pristine (B) APTES- (C) TSUA- (D) PTMO-coated SiO2 NPs. Black bars: bound protein, Grey bars: unbound protein. Coupled in pH 4 citrate buffer, 10 mM; pH 7.4 sodium phosphate buffer, 10 mM; pH 9 Tris buffer, 10 mM. Results of BCA protein assay expressed as µg mL-1 of protein in pellet (bound) or supernatant (unbound); Bet v 1 only as control; Graphs show mean ± SD of three experimental repeats. (E) Binding capacity in weight percent of four different SiO2 NPs with Bet v 1. Mean ± SD of three experimental repeats.
These results did not coincide with the predicted adsorption energies (Figure 1) suggesting that Bet v 1 possesses the highest binding affinity to APTES-coated SiO2 NPs. In contrast, in vitro studies revealed the lowest binding affinity to APTES-coated NPs among the four tested NPs. Additionally, the in silico modelling approach calculated almost similar binding affinities for all four NPs. This indicates that different surface characteristics, i.e. different ζ-potential values used for modelling, did not have a big impact on the calculated binding energies.
To investigate possible structural alterations or specific orientations of Bet v 1 upon non-covalent binding to the four differently coated NPs, an allergenic effector cell degranulation assay (huRBL assay) was performed. A specific orientation or conformational alterations of allergens like Bet v 1 could lead to destruction or inaccessibility of IgE epitopes, resulting potentially in crucial changes in the allergenic response against allergen-NP conjugates.
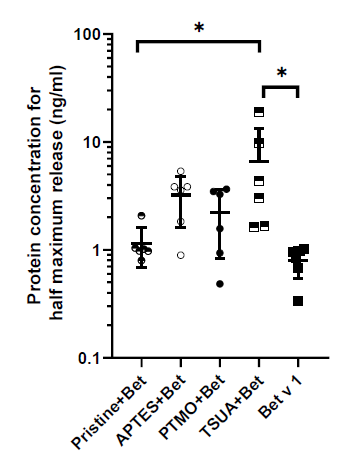 Figure 3. Basophil mediator release of allergic patient sera upon incubation with Bet v 1 only or allergen-NP conjugates. Protein concentration for half maximum release in ng mL-1 on log scale. Scatter plot shows mean ± SD of 6 different patient sera. Statistical analysis with GraphPad Prism 8; Ordinary one-way ANOVA followed by Tukey´s post hoc test for multiple comparisons; *p ≤ 0.05.
Figure 3. Basophil mediator release of allergic patient sera upon incubation with Bet v 1 only or allergen-NP conjugates. Protein concentration for half maximum release in ng mL-1 on log scale. Scatter plot shows mean ± SD of 6 different patient sera. Statistical analysis with GraphPad Prism 8; Ordinary one-way ANOVA followed by Tukey´s post hoc test for multiple comparisons; *p ≤ 0.05.
Figure 3 shows that there was a significant decrease in mediator release, i.e. less allergenic response, against Bet v 1 coupled to TSUA-coated SiO2 NPs. This observation may be due to either a change in the structure or a specific orientation of Bet v 1 impacting accessibility of the respective allergenic epitopes, when it was bound to this type of NPs.
Comparing these results with the results of the in silico modelling in Figure 1, it can be seen from the almost identical appearance of the heatmaps, that the modelling indicated that the preferred orientation of Bet v 1 was the same for binding to all four types of NPs. This contradicts with our in vitro investigations revealing one specifically preferred orientation of Bet v 1 coupled to one type of the four investigated NP types.
Discussion - strengths & weaknesses identified
The following gains were expected:
- The demonstration case sought to produce further experience with applying bio-nano interaction research that is less closely related to the so far conducted (eco-)toxicological experimental and in silico workflows.
- Since the work was mainly driven by PhD/MSc students, it was supposed to show if and when additional expert knowledge is needed. Based on the TA project, i.e. NC10, starting the work, the respective student received introductions to modelling and has been made familiar with the software and the input. This, as well as follow-up expert consultancy, was planned to be used to improve subsequent training material for the used services.
Ad 1. In regard to the first expected outcome, the goal was certainly achieved. The type of bio-nano interaction investigated here extended to a recombinant highly purified allergen preparation interacting non-covalently with four differently surface-functionalized silica NPs. Variations of bound protein to the different NP types were found as will be outlined in further detail below.
Ad 2. In contrast to the procedure conducted here, the UnitedAtoms in silico protein corona prediction tool is now available via the NanoCommons KB and can be run as an analysis service directly from there on predefined NPs. The students involved in this demonstration case served as beta testers of this newly integrated service and proved helpful for optimizing its functionality. A series of training sessions dedicated to the NanoCommons KB, i.e. NP definition, data/metadata upload, conduct of certain analysis tools, such as NanoXtract, UnitedAtoms, etc. has been forecast.
Identified strengths
The in silico protein corona prediction tool UnitedAtoms can be used offline, and in a high-throughput manner, as the user can queue up consecutive calculations of NP-protein interactions. Offline access requires a local computer, running the Linux operating system (OS), with sufficient computing power to perform these calculations together with an intermediate knowledge of Linux and the corresponding command line. To circumvent the requirements for a local machine running the calculations, the Transnational Access enables the use of remote machines running the UnitedAtoms software via a virtual network computing (VNC) access through a Secure Shell (SSH) network channel. While this mitigates some hurdles, the need for knowledge in Linux is still given, and it brings with it the need for technical knowledge to use the remote access as well as the addition of potential complications when accessing from an OS other than Linux. In this context, the integration of the in silico protein corona prediction tool into the NanoCommons Infrastructure, to be accessible directly from the NP entry in the NanoCommons KB, is a clear advantage, as much less computational skills are required from users. The user can choose any SiO2, TiO2, carbon and gold NP pre-defined in the KB and run a corona analysis directly from the KB. Currently, the database includes a library of potentials from atomistic modelling of NP-biomolecule fragment interactions for the above listed materials. For analysis, the NP-ID has to be selected in addition to the ID (Uniprot, PDB) of the desired protein for interaction. Then, the material of the NP has to be selected from the four different materials available and the specific size and zeta-potential values of the selected NP has to be entered. The tool calculates the adsorption energies for all possible orientations, outputs a map-file containing this information and generates a heatmap, which displays the adsorption energies of the corresponding angles phi and theta to depict the lowest adsorption energy and the corresponding degrees of rotation. This enables fast and comprehensive corona predictions with large data output in a very short timeframe and can be easily applied by inexperienced users.
Specific recommendations for further optimisations of in silico protein corona prediction
The attempted alignment of in vitro experimental analysis with the in silico protein corona predictions revealed substantial deviations between the results. Therefore, we aimed to identify the potentials and limitations of the current protein corona modelling tool and to list a number of recommendations. In summary, these recommendations comprise:
- To include environmental parameters, like pH value, into the simulations → Since our experimental results showed that environmental factors, like pH value and molarity, determine the mode and quality of interaction to a great extent, adequately simulating environmental conditions might be very important for the predictive power of the modelling tool. So far, the tool can address this only via simulating varying surface potentials of the NPs, however, altered surface potentials of the proteins, depending on the pH of the surrounding medium, are not taken into account. The upgrade of the tool using PROPKA 3 code evaluating the environment-dependent protein charges (Cedervall et al., 2007)(Deng et al., 2013) is currently underway.
- To consider compensating charges of neighbouring amino acids of folded proteins → Currently, the compensating charges are included in the model via an additive charge monopole model, i.e., if one residue is attracted to the surface via Coulomb force, another nearby may be repelled due to the opposite charge. What is not included is a higher order effect: match between a protein dipole and surface dipoles. Such a procedure would very unlikely be feasible for the corona model. It can be done using a detailed single-protein simulation but cannot be automated and scaled up to the entire corona.
- To include functional groups of NP coatings into calculations of potentials of mean force and hamaker constants → After NP coating, the proteins may much less interact with the core of the NP, but rather with the functional groups exposed on the surface. In vitro binding studies showed clear differences in the proteins´ binding behaviour depending on the differences in surface charge and hydrophobicity of the differently coated SiO2 NP types. In contrast, the in silico modelling results were almost identical for all types of SiO2 NPs. Therefore, it might be advisable to generate new potentials of mean force and Hamaker constants for the actual functional groups exposed on the NP surface and interacting with the 20 proteinogenic amino acids. At this time, however, an addition of an arbitrary chemical modifier to the model cannot be treated in an automated way. It requires a significant computational effort to parameterise the relevant interactions.
- To include the effects of hydrophobic interactions besides electrostatic and van der Waals forces → Our experimental results, in accordance with previous findings (Cedervall et al., 2007), revealed the importance of hydrophobic interactions on corona formation, since the TSUA-coated NPs, possessing a highly hydrophobic character, showed the highest binding capacity and altered epitope accessibility of adsorbed protein. While hydrophobic interactions are included in the model for the protein-NP core interactions, the additional effects originating from the surface modification are missing. Excluding this type of interactions per se, may limit the predictive power of the in silico modelling tool. If the development of novel NP descriptors, similar to recently developed protein descriptors like the GRAVY (grand average of hydropathy) (Duan et al., 2020), would be possible, it may become realistic to predict potential disruptions in the tertiary structure of the adsorbed proteins having implications on the biological outcome induced by hydrophobic interactions.
- To enable a direct assignment of predicted adsorption regions to the proteins 3D structure to facilitate the identification of possible regions of accessible epitopes for biologic reactions by experimentalists → The direct assignment of calculated adsorption energies and the corresponding angles to the respective PDB structure of the protein might be an optimisation potential for the data output. This would be very important for experimentalists, as it would facilitate identification of e.g. immunological epitopes and allow predictions of potential alterations in the biological outcome.
Conclusions on the overall achievement
A mature manuscript draft for an original article is available to be submitted to a scientific journal. It has the provisional title:
“Alignment of protein corona prediction with experimental analysis: showcasing an in vitro-in silico workflow providing FAIR data for nanosafety assessment of allergen-silica nanoconjugates”, co-authored by Ingrid Hasenkopf, Robert Mills-Goodlet, Litty Johnson, Mark Geppert, Ian Rouse, Vladimir Lobaskin, Dieter Maier, Albert Duschl, Iseult Lynch, and Martin Himly.
Based on the experiences from the demonstration case, PLUS researchers have adopted the strategy walked and, moreover, uploaded further raw data with corresponding metadata of work on self-produced NPs interacting with biomolecules to the NanoCommons KB. Presently, proposed new ontology terms for such novel types of bio-nano interactions have been inserted into the NanoSafetyCluster Terminology harmonizer developed in and provided by the GRACIOUS project, where new terms are collected and can be moderated by experts in the community, which has recently been established by NanoCommons in collaboration with the GRACIOUS project. When the discussion converged, accepted terms and their definitions have been included in the NanoSafety Cluster eNanoMapper ontology, maintained by NanoCommons for several years. The experimental-to-nanoinformatics workflow functioning in an integrated way through the NanoCommons KB, directly from the uploaded NP entry, was on display at the EuroNanoForum 2021 (see Figure 4), advertising the capacity of the NanoCommons Knowledge Infrastructure to industry and regulators, in particular for serving the Safety-and-Sustainability-by-Design concept, and at the Risk Governance session of the Nanosafety Training School on June 24, 2021.
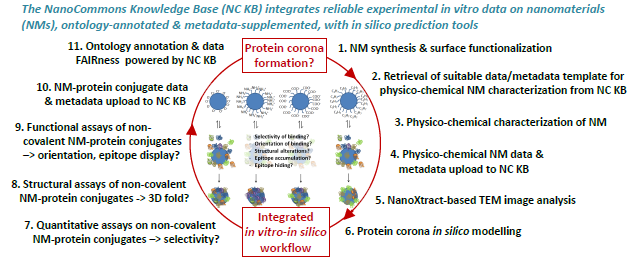 Figure 4. Central display item visualizing the experiment-to-informatics workflow of the poster presented in the Virtual Poster Session (Topic 3. Sustainable by Design: Embedding sustainability criteria throughout the life cycle of products and processes): “The NanoCommons Infrastructure – A Quick Guide to the WHAT and HOW of Experiment-to-Informatics Workflows Serving the Safe & Sustainable-by-Design Concept” by Martin Himly, Beatriz Alfaro Serrano, Dieter Maier, Thomas Exner, Iseult Lynch, and the NanoCommons consortium.
Figure 4. Central display item visualizing the experiment-to-informatics workflow of the poster presented in the Virtual Poster Session (Topic 3. Sustainable by Design: Embedding sustainability criteria throughout the life cycle of products and processes): “The NanoCommons Infrastructure – A Quick Guide to the WHAT and HOW of Experiment-to-Informatics Workflows Serving the Safe & Sustainable-by-Design Concept” by Martin Himly, Beatriz Alfaro Serrano, Dieter Maier, Thomas Exner, Iseult Lynch, and the NanoCommons consortium.
Generalizing from the experiences in this demonstration case, we learned that the initial TA service to learn to apply the UnitedAtoms protein corona prediction tool at UCD plus support thereafter was an excellent opportunity for early-stage researchers to get it running, even remotely via the established VNC and SSH access. Upon integration of the UA tool into the Knowledge Infrastructure, rendering it being accessible directly from the nanoparticle entry in the NanoCommons KB, the required TA time and resources will be further minimized, however, it needs to be warranted that the most recent version of tools are functionable to provide most reliable modelling results.
References
- Cedervall et al., 2007: Cedervall, T.; Lynch, I.; Lindman, S.; Berggård, T.; Thulin, E.; Nilsson, H.; Dawson, K. A.; Linse, S. Understanding the Nanoparticle–Protein Corona Using Methods to Quantify Exchange Rates and Affinities of Proteins for Nanoparticles. PNAS 2007, 104 (7), 2050–2055. https://doi.org/10.1073/pnas.0608582104.
- Deng et al., 2013: Deng, Z. J.; Liang, M.; Toth, I.; Monteiro, M.; Minchin, R. F. Plasma Protein Binding of Positively and Negatively Charged Polymer-Coated Gold Nanoparticles Elicits Different Biological Responses. Nanotoxicology 2012, 7 (3), 314–322. https://doi.org/10.3109/17435390.2012.655342.
- Duan et al., 2020: Duan, Y.; Coreas, R.; Liu, Y.; Bitounis, D.; Zhang, Z.; Parviz, D.; Strano, M.; Demokritou, P.; Zhong, W. Prediction of Protein Corona on Nanomaterials by Machine Learning Using Novel Descriptors. NanoImpact 2020, 17, 100207. https://doi.org/10.1016/j.impact.2020.100207.
- Lahiri et al., 1999: Lahiri, J.; Isaacs, L.; Tien, J.; Whitesides, G. M. A Strategy for the Generation of Surfaces Presenting Ligands for Studies of Binding Based on an Active Ester as a Common Reactive Intermediate: A Surface Plasmon Resonance Study. Anal. Chem. 1999, 71 (4), 777–790. https://doi.org/10.1021/ac980959t.
- Power et al. 2017: Power, D.; Rouse, I.; Poggio, S.; Brandt, E.; Lopez, H.; Lyubartsev, A.; Lobaskin, V. A Multiscale Model of Protein Adsorption on a Nanoparticle Surface. Modelling Simul. Mater. Sci. Eng. 2019, 27 (8), 084003. https://doi.org/10.1088/1361-651X/ab3b6e.

