Generation and upload of individual datasets
Preparing specific datasets from single users (e.g. project consortia, research groups), including rich, semantically annotated metadata characterising the experimental procedure of data creation and improving the interpretability and comparability of the data, needs specific workflows to be followed to ensure proper upload to the NanoCommons Data Warehouse (DW) and subsequent integration into the NanoCommons Knowledge Base (KB) as well as to assess the quality and FAIRness levels of the data to enable inclusion of these parameters with the dataset.
These have to be aligned and harmonised with the workflows for integration of complete databases and, thus, similar information have to be provided before the dataset preparation and uploading starts. The key data required from the data owners, presented schematically in Figure 1, include:
- Detailed information of the person submitting the data along with contact details;
- publication status and whether a DOI already exists;
- an abstract describing the field of study, scope and endpoints of the experiment;
- the dataset format;
- existing metadata based on the requirements either of a specific data warehouse, including the NanoCommons DW, or on the general requirements of the KB, if no appropriate DW exists;
- data and metadata annotation to the NanoCommons (eNanoMapper) ontology;
- an evaluation from the submitter on the dataset quality based on metrics developed by the NanoCommons consortium;
- the desired FAIRness level, Openness and license assigned based on what is available through the NanoCommons KB.
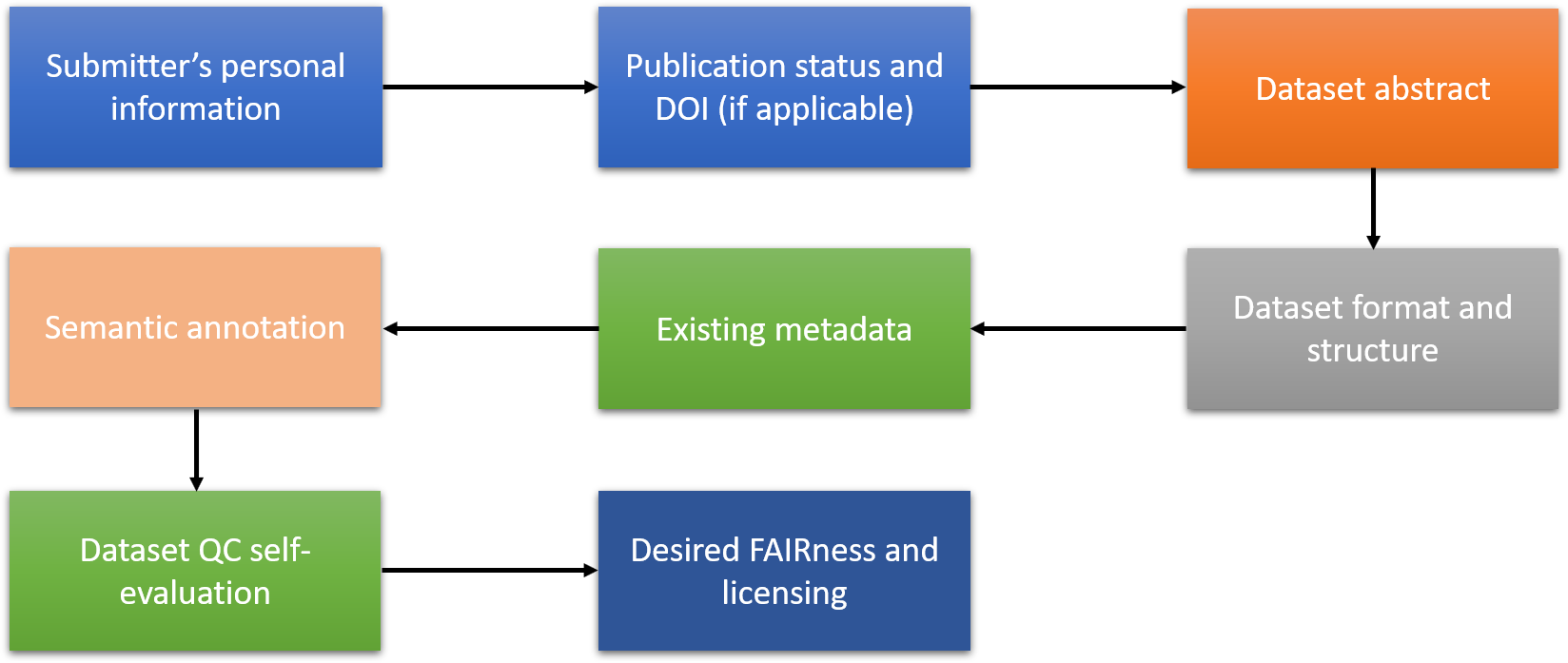 Figure 1. NanoCommons workflow for the implementation of datasets into the KB and DW.
Figure 1. NanoCommons workflow for the implementation of datasets into the KB and DW.
Following the above processes and keeping them aligned with workflows for complete databases is critical to ensuring a streamlined and straightforward process, with less bugs (or errors).
Data Preparation Guidelines adopted in NanoCommons
Data are valuable resources that can be used and reused by the scientific community. The data integration and sharing functionality provided by NanoCommons facilitates new scientific inquiry, avoids duplication of experiments and data collection, and provides rich real-life resources for method validation, risk assessment of NMs, as well as education and training. However, this is only possible if the data is generated and provided in a harmonised and interoperable way, annotated by rich metadata, and is produced following state-of-the-art quality standards. In the “Managing and Sharing Data” guide by the UK Data Archive (Van den Eynden et al., 2014) this is emphasised in the following way: “A crucial part of making data user-friendly, shareable and with long lasting usability is to ensure they can be understood and interpreted by any user. This requires clear and detailed data description, annotation and contextual information”.
Data curation sheets provided for the uploading of data to specific databases or developed through the NanoCommons project have been created considering data and metadata quality standards, and a core aspect of NanoCommons dissemination and training activities will be to promote the use of these broadly by the nanosafety community. Such standards and good data management practices should be at the heart of all credible science and should be kept in mind during the evaluation and refinement of curation sheets, the development of new data templates and during the complete processes supporting the integration of Users data into the NanoCommons KB.
Following the guideline and checklists promoted by NanoCommons and outlined in this report, will help the TA and independent users to:
- know the legal, ethical and other obligations regarding research data, for each of the research participants, colleagues, research funders and institutions;
- implement good practices in their experimental design and data management across the whole data life cycle in a consistent manner;
- assign roles and responsibilities to relevant parties in the research;
- design data management responsibilities and processes according to the needs and purpose of research;
- incorporate data management measures as an integral part of their research cycle;
- implement data integrity and quality risk management checks and processes;
- anticipate difficulties in making data available and design measures to overcome these; and
- implement and review data management practices throughout the research process as part of research progression and review.
As part of the activities of NanoCommons, we are also developing a certification in Nanosafety data curation. All NanoCommons checklists are linked to existing community consensus in best practice for reporting data, including the recently published Minimum information reporting in bio–nano experimental literature (MIRIBEL)(Faria et al., 2018) suggestions, which provides a starting point for the community to build upon, and other data reporting standards.
Key Concepts for Data Curation
To allow data usage without the need to refer to external sources like publications, final reports, working papers, or the Electronic notebooks (ELN) used to generate the data, all important information should be included in the data submission. On the data completeness and quality page of this handbook, the problems of how to define and evaluate data completeness and quality are addressed in great detail and different understandings are presented.
In the context of data curation, not only the quality of the original experimental data needs to be considered but also quality considerations associated with curated data. Quality considerations associated with curation include the probability of transcription errors and possibly whether a given dataset, structured according to some standardised format (e.g. XML based), was compliant with the rules of the applicable standardised format (e.g. as documented via an XML schema). Such compliance, amongst other aspects of data quality, could be determined using validation software.
Dataset integration
NanoCommons offers, besides database integration and accessibility, services to single users (defined as project consortia, research groups, individuals etc.) to prepare, curate and submit datasets that are not already included in other linked databases. For these datasets to be integrated, the workflow described here ensures that all information required for the NanoCommons DW or another integrated database are provided and that all NanoCommons quality standards are kept. The entry point for data submission is through the NanoCommons website and via a members’ area page. To access the members area, registration is required from the interested parties during which certain personal data (protected under the GDPR law (as per the NanoCommons GDPR statement) and not shareable with third parties) will be collected. While the entry point is via the NanoCommons website the main site is hosted under the Biomax’s platform, to which the user is redirected and where the rest of the needed information is collected. The information, which will be required for dataset(s) integration in the DW, is listed in the following sections. Note that for Users applying via TA to NanoCommons the data from their TA application form can be transferred across automatically.
Personal information
The personal information that will be requested from single users will be:
- Submitter’s name and email
- Submitter’s affiliation and group (academia, SMEs, industry, regulatory)
- Owner Group (whether the dataset(s) belong to individuals, research groups, EU, national or international projects etc.)
- Field of expertise applied to the dataset
- Acceptance of the terms and conditions and ethics requirements for submitting, accessing and using data through the NanoCommons DW.
Following registration, the application will be tested, verified (via email) and approved automatically by the system and the registrant will be redirected to the NanoCommons KB for data submission. There, specific metadata and data information will be required that will also act as an initial measurement of the submitted dataset’s quality. The information ideally will be provided using a template provided by the system based on a standard curation format that will be structured and harmonised to fit the DW requirements. Otherwise, the option to fill in the details online and to submit a separate dataset may be provided, which may be easier for users.
General dataset details
Following the submission of the submitter’s personal details, general bibliographic information about the data will be required:
- Dataset owner (individual, research group, institution, EU, national or international project)
- Names, and affiliations of the experimental team (one name for each experimental end-point)
- Name and contact details of the person to act as a scientific contact point
- Application (physicochemical properties, hazard, exposure and fate, etc.) and toxicology (human heath, environmental, etc.) domains relevant to the dataset
- Origin of data (raw, summary, computational, published) - ideally several of these will be provided together
- Publication status (published, open access, under preparation, unpublished etc.) and relevant details
- Assigned DOI or any other unique ID for the dataset. Where no DOI is present the option to acquire a NanoCommons DOI via Zenodo will be provided for the dataset.
- Desired FAIRness and license type assigned or to be assigned to the submitted dataset, based on the tiered system developed within NanoCommons
- A short summary (abstract) describing the field of study, scope and endpoints of the experiments included in the dataset.
Based on the submitted information, the NanoCommons DW will check and identify existing relevant databases for the data to be uploaded to. In any case, the user will upload their datasets in the NanoCommons DW, to simplify the process, and if necessary, the NanoCommons data management system will redirect the data to the most appropriate database.
Experimental details and metadata
The next step in the process will be to gather specific details on the dataset that will allow successful integration into the NanoCommons KB (or the alternative platform identified) and potential enrichment and annotation. These will include:
- The dataset format and whether is human or machine readable (e.g. Excel template, HTML, XML JSON)
- Experimental metadata, which include methods, protocols/SOPs, best practice guidelines utilised for reporting (if any) such as MIRIBEL, instruments used and the format used to submit the metadata (e.g. extracted from ELNs, use of ACEnano questionnaires, instance-like, free text)
- Whether the data and metadata have been annotated or not and whether mapping of the ontological codes used (if it hasn’t been mapped, the user will be directed to the NanoCommons ontology tools implemented in the NanoCommons KB and will be guided through the process of ontologically mapping their dataset).
- Existing data enrichment and whether the user desires to use the enrichment functionalities offered by the NanoCommons KB.
Data governance
Finally, the submitter will be required to fill in a short survey to act as an initial QC of the submitted dataset. This questionnaire will cover several aspects of the submitted data and metadata. The required information will include:
- any data cleansing approaches applied;
- any existing data gaps;
- protocols and methods origin and whether these are standardised (e.g. ISO);
- errors and uncertainties reporting;
- statistical methods applied (if applicable) and relevant tests applied to ensure test validity (e.g. linearity, normality, dataset size); and
- any ethics statements and/or ethical approvals, where applicable, for data submitted from human and/or animal in vivo and in vitro studies.
Dataset preparation and curation
All the information provided by the user will then be evaluated by the NanoCommons team, beginning with a quality evaluation. For example, in cases where substantial amounts of information/data/metadata is missing, the expert panel responsible for judging the quality of the dataset submitted will decide whether to accept it or not, and if it does accept the dataset will assign appropriate flags regarding the completeness and data quality. The panel will also decide if the dataset type fits any of the existing databases integrated in the NanoCommons KB. This will help avoid further data fragmentation by establishing publicly available data repositories, in terms of both access and submission of datasets, for physicochemical (e.g. physicochemical properties and next generation NMs), biophysical (e.g. corona composition), human hazard, environmental fate and hazard, (product) handling, release and exposure and modelling nanosafety data. If a suitable database is identified, the contact to the data manager will be established and the technical infrastructure (login account, access rights, etc.) will be set up to allow the data to be uploaded. Since most of the considered databases are provided and managed by different NanoSafety Cluster or international projects, this administrative work has to be performed separately for each database. With increasing integration of the resources over the duration of the NanoCommons project, this process will be substituted by a single-sign-in option allowing uploading and access to all databases using one single account, but still providing features to assign different access rights. The user will then be asked to prepare the dataset according to the requirements of the selected database using the provided data curation templates or upload functionalities of the database. This step will also include the semantic annotation of the data and metadata following the database-specific rules and NanoCommons recommendations. These tasks will be strongly supported training activities and, during the runtime of NanoCommons, the TA offerings.
In case no suitable database is identified or it is not available for submissions from the “public”, the NanoCommons DW will provide the hosting of the datasets. If the datasets are coming from larger consortia, and many sets of the same type and format are expected, specific data curation templates will be designed or even specific upload functionality for the DW will be developed within the TA actions (i.e. the user will be directed to apply for TA funding to support the development of bespoke tools for their datasets). This will be done through intensive collaboration between the user and the NanoCommons team and will take all the guidelines and checklists described here into account. The new templates will be aligned, harmonised and structured around existing concepts to facilitate the interoperability with the other NanoCommons data sources. Semantic annotation of the data model (data schema), as well as the data and metadata using the concepts developed jointly by the NanoCommons and OpenRiskNet projects and the ontologies endorsed by these will be enforced to make the datasets seamlessly integrated into the semantic model of the NanoCommons interoperability layer. The templates will also be shared with other relevant projects to facilitate their data upload also, thereby accelerating the integration of the currently disparate datasets arising from smaller-scale EU and national projects, such as for example Marie Curie ITNs, Individual Fellowships etc. A list of relevant national projects has already been compiled as part of D10.2, and this will be further updated over the coming months. The range of projects targeted as part of the TA dissemination activities will include those with a more nanomedicine or environmental applications of NMs focus (e.g. for environmental remediation, nanopesticides etc.), as many of the core issues of NMs characterisation and safety assessment will be common across fields.
References
- Faria et al., 2018: Faria, M.; Björnmalm, M.; Thurecht, K. J.; Kent, S. J.; Parton, R. G.; Kavallaris, M.; Johnston, A. P. R.; Gooding, J. J.; Corrie, S. R.; Boyd, B. J.; Thordarson, P.; Whittaker, A. K.; Stevens, M. M.; Prestidge, C. A.; Porter, C. J. H.; Parak, W. J.; Davis, T. P.; Crampin, E. J.; Caruso, F. Minimum Information Reporting in Bio–Nano Experimental Literature. Nature Nanotech 2018, 13 (9), 777–785. https://doi.org/10.1038/s41565-018-0246-4.
- Van den Eynden et al., 2014: Eynden, V. van den; UK Data Archive. Managing and Sharing Data: A Best Practice Guide for Researchers; UK Data Archive: Colchester, 2011. http://www.data-archive.ac.uk/media/2894/managingsharing.pdf.
Appendix - The NanoCommons “User data” checklist
- Submitter’s personal information:
- Submitter’s name and email
- Submitter’s affiliation and group (academia, SMEs, industry, regulatory)
- Owner Group (whether the dataset(s) belong to individuals, research groups, EU, national or international projects etc.)
- Field of expertise applied to the dataset
- Acceptance of the terms and conditions and ethics requirements for submitting, accessing and using data through the NanoCommons DW.
- General dataset details:
- Dataset owner (individual, research group, institution, EU, national or international project)
- Names, and affiliations of the experimental team (one name for each experimental end-point)
- Name and contact details of the person to act as a scientific contact point
- Application (physicochemical properties, hazard, exposure and fate, etc.) and toxicology (human heath, environmental, etc.) domains relevant to the dataset
- Origin of data (raw, summary, computational, published) - ideally several of these will be provided together
- Publication status (published, open access, under preparation, unpublished etc.) and relevant details
- Assigned DOI or any other unique ID for the dataset. Where no DOI is present the option to acquire a NanoCommons DOI via Zenodo will be provided for the dataset.
- Desired FAIRness and license type assigned or to be assigned to the submitted dataset, based on the tiered system developed within NanoCommons
- A short summary (abstract) describing the field of study, scope and endpoints of the experiments included in the dataset.
- Experimental details and metadata:
- The dataset format and whether is human or machine readable (e.g. Excel template, HTML, XML JSON)
- Experimental metadata, which include methods, protocols/SOPs, best practice guidelines utilised for reporting (if any) such as MIRIBEL, instruments used and the format used to submit the metadata (e.g. extracted from ELNs, use of ACEnano questionnaires, instance-like, free text)
- Whether the data and metadata have been annotated or not and mapping of the ontological codes used (if it hasn’t been mapped, the user will be directed to the NanoCommons ontology tools implemented in the NanoCommons KB and will be guided through the process of ontologically mapping their dataset).
- Existing data enrichment and whether the user desires to use the enrichment functionalities offered by the NanoCommons KB.
- Data governance:
- Any data cleansing approaches applied;
- any existing data gaps;
- protocols and methods origin and whether these are standardised (e.g. ISO);
- errors and uncertainties reporting;
- statistical methods applied (if applicable) and relevant tests applied to ensure test validity (e.g. linearity, normality, dataset size); and
- any ethics statements and/or ethical approvals, where applicable, for data submitted from human and/or animal in vivo and in vitro studies.