NanoCommons Demonstration Case - Data management concept for ecotoxicological information on potential environmental release of ENMs as derived from the NanoFASE project
This case demonstrates how data management can be organised in a large multidisciplinary project. This case has been finalised.
Table of contents
Background and aims and contribution of NanoCommons partners
The H2020 project NanoFASE generated complex, multi-level data from all environmental compartments, covering the entirety of the NMs’ life cycle. The project’s experiment work included different complexity levels, starting from simple NMs synthesis and characterisation to highly complex outdoor experimental systems (mesocosm) that examined environmental fate and behaviour of NMs under controlled conditions to understand the functional and behaviour patterns of NMs in exposure-relevant environments. This data complexity and range provided challenges to standard data and metadata capturing, curation, QC and required the ability to connect multiple levels of information such as the characterisation of the pristine NMs and the environment, the specific changes to NM due to aging in the specific environment, and the effect of the aged NMs on the respective environmental compartment.
The main objective of this case study was to establish a management workflow for the data-related processes at a large multidisciplinary project level, which includes the data preparation needed, the steps to introduce these processes and their benefit to the entire consortium, the establishment of communication and trust with the different partners and the training needed to familiarise the partners with the data management processes and their application. Subsequently, the aim was to integrate the entirety of the NanoFASE data generated by multiple partners and make it interoperable with the NanoCommons Knowledge Base and other similar databases, like the US-equivalent CEINT-NIKC.
To achieve this, UoB and BIOMAX developed and refined a data structure and format (expanding the original CEINT-NIKC format) to be able to capture all data types, fit the needs of the NanoFASE consortium and is integratable with the NanoCommons e-infrastructure. To integrate the NanoFASE data generated by multiple partners, and make it interoperable we modified the existing CEINT-NIKC template in collaboration with partners UK-CEH and Duke University, who were the first to develop it. The process included the simplification, as much as possible, of the NIKC template, while ensuring its interoperability with the original. Based on this work, we developed in collaboration with all involved partners short visual and longer text-based guides on how to fill these complex templates. The produced datasets were integrated into the H2020 NanoCommons infrastructure, following development of the semantic model, the data capture templates and the graphical visualisation of the experimental steps, facilitating API based access and tool integration to interrogate and model the data, via NanoCommons, H2020 NanoSolveIT and beyond.
Results
The case provided invaluable insights on the data management processes required in the case of a large research project, where different types of experimental and computational data have been produced. The lessons-learned included the optimum approach regarding managing data from different work packages and specific partners, the potential types of data and metadata, the needed data templates and the integration of these data into the NanoCommons Knowledge Base.
NanoFASE contained 35 partners, spanning the entire data life cycle and data roles as described Papadiamantis et al. (Papadiamantis et al., 2020). The partners included representatives from all major stakeholders, i.e. academia, industry, regulatory researchers. As a result, the data produced needed to be handled and processed using different approaches, so as to respect potential commercial and regulatory sensitive data.
Based on the work performed, a clear set of challenges was identified with respect to implementing a clearly defined data management process. Firstly, substantial information communicated using simple language was required, focussing to a large degree on the benefits for the data creators. This was necessary to trigger the required mental change for the need to shift from established processes to more modern and digitised data management. This was achieved using a set of consortium-wide workshops, where the benefits and responsibilities of modern data management processes were presented and discussed. During this time the significance of the project’s coordination team became evident, as the strong support from the NanoFASE coordinator played a key role in consortium members surpassing their reservations. The key arguments that seem to convince the consortium members were the added value to the data produced from the creators via data interoperability and combination and the potential for increased number of citations originating from applying Digital Object Identifiers (DOIs) to the datasets and publishing via an established database. Another benefit was the fact that users had the ability to use the curated and structured datasets in support of their publications, as most of the peer-reviewed journals require submitting the data along with any manuscript.
Substantial obstacles in acceptance were based on technical details, which were hard to be apprehended in full from researchers lacking relevant knowledge. This was also the case for metadata standards and completeness that were required to be reported, as the data creators were unaware of their usage in terms of interoperability and their significance for other data roles. This made evident the need for implementing data shepherd roles, a new role define in Papadiamantis et al. (Papadiamantis et al., 2020) partly based on the experiences in this demonstration case, in large and/or multidisciplinary projects that would be able to communicate in a simple and clear way the needs and requirements of all data roles. Another substantial obstacle was encountered during data annotation. In many cases, specific terms were used under a clearly defined, very field-specific context (e.g. the term deposition was defined as the drop of water or liquid on the ground). This created issues in the wider project framework. To overcome this, respective workshops were organised during which terminology was gathered and potential conflicts were identified. In these cases, discussion was performed with the data creators and a solution using a set of terms combination was usually proposed. Where no consensus could be reached the decision on how to address this was taken at a higher project level, taking into account the need for data harmonisation across and beyond the project.
These workshops were followed by data filling workshops using the NIKC template, so that the NanoFASE data could be interoperable with respective data curated by the Center for the Environmental Implications of Nanomaterials (CEINT), Duke University, USA and the European Centre Research And Teaching In Geosciences De L’envi (CEREGE), France. During this phase it became evident that the use of joint data curation workshops was useful for users to understand the basics of such a complex template, but partner-specific sessions were required to fully understand the methodology and process for filling in the required data and metadata template sections. As a result, a set of personalised sessions were organised with partners to help them design and fill in their templates, which was followed by continuous support to resolve any issues and answer any questions that arose.
A characteristic example of applying the above process, was the capturing of complex NanoFASE mesocosm data along with all relevant metadata into a technical system that ensures FAIR availability. Each dataset now is findable by unique, stable identifier and rich metadata and, via registration of the NanoCommons Knowledge Base, is indexed at re3data.org and fairsharing.org. The data is accessible by user interface and technical access protocols (API, Schema) and interoperable due to the common language based semantic concepts and mappings to ontologies as well as support for the standard ISA-tab format. Finally reusability is supported by a template data analysis agreement and, at the end of the embargo period, each dataset will be associated with a clear license which is supported by the governance and provenance functions provided by the NanoCommons Knowledge Base.
As a result of these efforts by all data generating partners within NanoFASE, and the dedicated data shepherd role played by UoB and the technical support provided by Biomax, the publicly accessible NanoCommons Knowledge Base now contains and provides access to over 5,400 measured instances of ENMs collected from different environments (air, water, soil, sediment, waste treatment) at different ages and assessed and characterised with a multitude of methods. This rich data collection is available by user and application programming interfaces to inform nanoinformatics applications such as read across, computational modelling, machine learning or nanosafety and risk assessment.
In collaboration between UoB, BIOMAX and the partners providing mesocosm data a data capture template was developed which made sure that data is accompanied by and correctly linked to the necessary metadata (bibliographical, partner, experimentalists) and experimental (protocols, methods, variable definition) required to ensure findability and re-use. The template was implemented as an Excel file with eight separate tables capturing publications, institutions, people, ENM instances, measurements, protocols, instruments and variable dictionary (Figure 1).
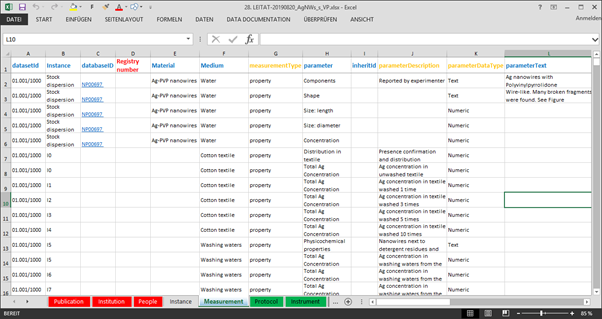 Figure 1. NanoFASE data capture template.
Figure 1. NanoFASE data capture template.
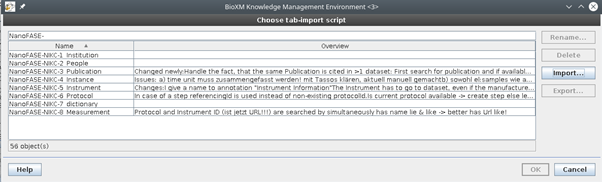 Figure 2. Example of one of the eight semantic mapping agents implemented.
Figure 2. Example of one of the eight semantic mapping agents implemented.
Correspondingly we developed eight semantic mapping agents (Figure 2 and Figure 3) which take the different types of information from each of the Excel tables and map them to the concepts in the semantic model. These mappings connect the different levels of information such as institution, measured ENM instance or measurement protocol within one data-set and at the same time connect them to all other existing datasets (Figure 4). In this way one can, for example, search for all datasets measured with the same type of instrument.
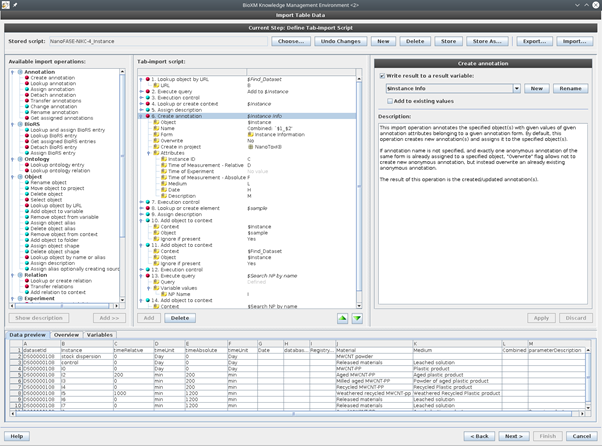 Figure 3. Semantic mapping agent for instance information. Depicted are the metadata assignment step and the sample – instance mapping step.
Figure 3. Semantic mapping agent for instance information. Depicted are the metadata assignment step and the sample – instance mapping step.
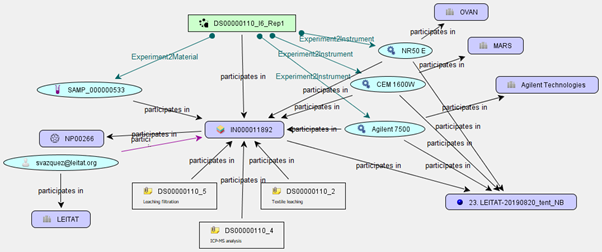 Figure 4. A specific instance (box in centre) mapped to particle characterisation measurements (green box on top) and associated metadata such as the ENM name and composition, data owner, protocols, instruments and dataset.
Figure 4. A specific instance (box in centre) mapped to particle characterisation measurements (green box on top) and associated metadata such as the ENM name and composition, data owner, protocols, instruments and dataset.
Several different access routes are provided for convenience in order to allow users to start their experience / data management journey according to their personal preferences or initial available information, for example depending on whether they are visual (e.g. flow chart and the visual representation of the experiment) or most list-based in their thinking. Overall, from any access point all information is interlinked and therefore available.
The interlinked information can then be accessed by starting from the dataset, from the measurement, from the protocols, or the ENMs, and other queries are being added as modelling tools are implemented. Currently most datasets are linked to one specific publication, and can also be searched via publications. However, as more and more data becomes re-analysed in different contexts this may change and is already accommodated within the semantic model.
For each dataset a number of ENM instances can exist as becomes visible if listing data by instances (Figure 5). Data itself is always associated with a Measurement parameter. Therefore, another way to access information is to select a specific measurement parameter, e.g. diameter, and list all available measurements (Figure 6). Finally, data can be accessed by either protocol or instrument used during its production. Both routes link to the corresponding datasets and from there to the measured data.
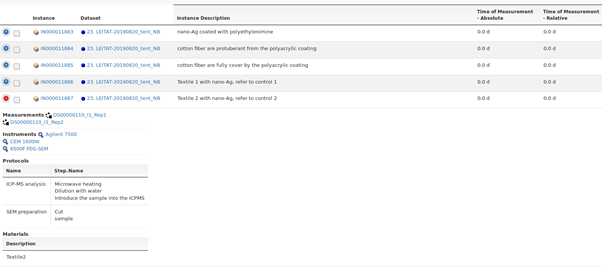 Figure 5. List of Instances linked to datasets, measurements, instruments, protocols and materials.
Figure 5. List of Instances linked to datasets, measurements, instruments, protocols and materials.
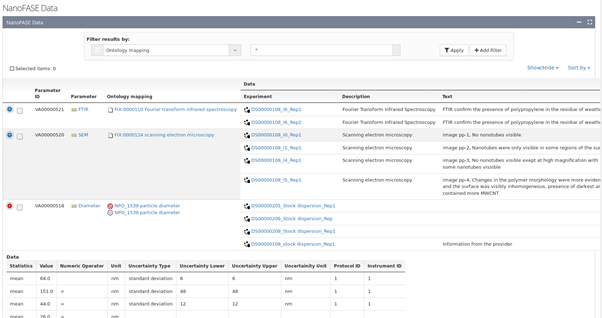 Figure 6. List of measurement parameters, linked to ontology terms, measurements and as an example visualising data for one specific parameter.
Figure 6. List of measurement parameters, linked to ontology terms, measurements and as an example visualising data for one specific parameter.
Early efforts to map computational data from NanoFASE to MODA templates were also undertaken, and will feed into the cross-WP MODA structuring and ontology mapping activities described in D2.8 on the work plan for the final phase of the NanoCommons project.
Discussion - strengths & weaknesses identified
This analysis of the successes and remaining weaknesses of the implemented data curation and management approach coordinated and heavily supported by the data shepherd negotiating between all roles is presented here in two parts. First, the data shepherd perspective will be given followed by the testimony of two data providers.
Data shepherd perspective
Data management needs to start very early in project - despite the best of intentions in NanoFASE, by the time the project started seriously working on the data management, most of the experimental data had already been generated, and thus it was a larger effort around organization and curation of the data as generated rather than a pre-established set of data capture templates into which the raw data could be processed as it was generated. Workflows were mostly in people’s heads, as loose documents describing the main timings but not necessarily all the intermediate steps which are often “assumed” (e.g., NMs exposure would imply that NMs were first dispersed to the required concentration in the specific medium which was prepared via a specific recipe following a specific protocol and tested to ensure compliance with the specifications in terms of pH and conductivity, for example). Initial efforts to collect and collate protocols were not as effective as they could have been, in part due to the fact that large parts of the project were around optimising the protocols, so final versions only came at the end of the project. Similarly, some protocols were word documents, some excel sheets, and were not linked in a structured fashion, so all this needed to be worked through with each partner on a case-by-case basis to understand their experimental workflow. In fact, the optimum approach identified was that an initial series of consortium-wide workshops are required to explain to participants the philosophy of the data management process, their responsibilities and above all the benefits coming out for them. These workshops, performed during the project’s General Assemblies helped with “changing the mentality” of the partners and introducing them to the benefits of structured data management. These were followed by joint hands-on workshops on the data templates (MODA, NIKC etc.), the types of metadata, semantic annotation etc. While useful, these workshops demonstrated the need for targeted workshops with each partner separately, as there was strong difficulty from partners to work with generic datasets and apprehend the data curation process. Needs for a very hands-on approach initially - data “shepherding”: The overall level of familiarity of consortium members with data management approaches was very low initially. Early consortium meetings focussed on introducing some of the key principles and approaches, and raising awareness, and later became more hands-on in terms of having the researchers work on their individual datasets to organise, curate, ontologically map, and graphically map their datasets, as well as collecting the relevant metadata including instrument details etc. (which actually becomes data itself also allowing uses of the NanoCommons KnowledgeBase to search for data generated using a specific assay or method). Face to face workshops and dedicated time working with the individual partners was needed - i.e. shepherding the experimentalists through the various data management steps, reassuring them that they were “doing it right” and steering them if they got confused. Compliance and engagement varied initially, with some partners (albeit with more straightforward experimental systems) “getting” it straight away while others, typically those with complex experiments and multiple factors being measured in parallel related to the ENM, the surroundings and the test organisms, struggled a little more to know best how to describe their experiments and thus their data capture template needs. Often several iterations of the data capture template / visual representation of the experiment were needed as through discussion with the data shepherd it emerged that “oh yes we do measure solution pH and dissolved oxygen at regular intervals”, or “oh yes there is a specific protocol for how we extract the soil pore water” etc. Our NanoCommons data shepherd (Tassos Papadiamantis) also spent several days with each NanoFASE partner working hands-on with them on their datasets. A lot of the lessons from these sessions have been distilled out into the user guidance co-developed with NanoFASE, wherein the data capture template used by NanoFASE is presented with instructions in the form of two tutorials: one is a visual/graphical manual, and the second is a text-based manual that contains all the analytical details of the process.
Also, when working through the process again with newly started projects ASINA and SabyNa, the possibility to embed the data management steps from the outset was invaluable. In their cases, the lessons learned from this case provided a clear map on the best data management processes. These started to be implemented very early in the projects’ lifetime, with the assignment of within-project data shepherds that are in continuous contact with the NanoCommons team. Furthermore, consortia meetings took place, as soon as the respective TAs were launched, to inform the data creators of their responsibilities and benefits. The in-house data shepherd commenced and gathered all types data and metadata that were going to be produced by each partner and designed the data workflows and capturing templates in collaboration with the data creators. During this phase, the NanoCommons team provided advice on the data workflows development, as well as on technical details in preparation of partner-specific meetings. Following this, the NanoCommons team supports the ontological annotation of the data workflows, including guidance on new term definition.
Strong push from the project coordinator was needed: NanoFASE was the first major nanosafety project to attempt to have 100% of its data curated and available for re-use after the project embargo period (September 2021). To achieve this, buy-in at every level was required - Coordinator, WP leaders, Task leaders, partner lead scientists and PhD and postdoctoral researchers. To generate and maintain this buy-in, dedicated hands-on sessions at every 6-monthly consortium meeting were required, initially focussing on the key concepts and later moving towards hands-on data curation sessions. Without this push, and the threat of withholding monies from the final payment if data was not in the KnowledgeBase, this level of data management could not have been achieved. However, we would hope that in subsequent projects the effort would be lower, in part as researchers were already familiar with the concepts, and in part because a lot of the hard work is the initial investment in establishing the templates and the visual representation of the experiments, and collection of the metadata, much of which can be reused or tweaked for subsequent projects / experiments. Thus, a key measure of success is assessment of whether or not the data management practices established in NanoFASE are embedded into the working practices of the researchers in the partner institutes.
Large Initial investment of time and effort needed, but once established should be easier to maintain these good practices: As a project, NanoFASE invested a significant amount of time into engaging with the consortium in order to introduce and encourage adoption of these approaches, initial training, and workshops to facilitate implementation. This effort included approximately 300 person hours involved in two in-person workshops, a further three virtual workshops, as well as approximately Y hours of Data Shepherd time. These efforts resulted in a total of around 90 comprehensive datasets, including several experimental steps.
Data provider and curator perspective
We attended the initial meetings/presentations outlining the principles of the data management system. Though the principals were understood, the workshop organised for several groups that involved data-heavy mesocosm experiments was particularly useful to identify the template outline, different needs associated with each partner’s data sets, organism and analysis type. These discussions aided the implementation for our individual experiments. However, we also spent an afternoon working with Tassos Papadiamantis, as the data shepherd, working hands-on with our group’s specific experimental data templates. This ability to ask questions relating specifically to our work and finding out how best to capture the nuances between different experiments was the real crystallisation point for us. After this session, we were able to populate the templates and felt more confident creating and modifying templates, we also felt able to help others who also needed to perform the same tasks. The population of the templates took between 2-3 hours per experiment, including putting the protocol in the correct format.
For all our NanoFASE experiments, we were entering data after the experiments had finished and the following is written with this in mind. From a lab-users perspective, we feel that the template lacked some human “reader friendly” components which, we felt, would have made the template population process quicker. Furthermore, we feel such components would not only make the data enterer more confident but would also have made the template more usable as a means of collecting data from the lab and for accessing the data in-house. Our own data organisation is naturally grouped in separate files by data type. The template contains many empty cells, this meant that we needed to filter rows by the parameter column to input blocks of the same data type into multiple instances. However, while filtering one column, the limited number of cells filled for other columns (such as the material column), made it more difficult to interpret whether data was being inserted into the correct rows. We also found that the lack of experimental information in the instance column made the population process slower. In such a large document, the data enterer was required to continually double check they were entering data into the correct rows. This, in our experience, meant flicking back and forth from the instance tab containing some of the experimental information relating to the identifier. It was felt that more information columns in the instance tab may also make it easier to keep track of the document. In later attempts to populate templates, we filled empty cells and added information columns on the measurement tab, such as adding instance information, this made the process much quicker and also aided the final confirmation stage when looking for errors. We then cleared the cells post-population. Without the ability to extract data rows in a usable form, we are less likely to use the templates, as they currently are, for in-house use and retrieval of data.
However, this experience with the NanoFASE data did help us to think about the way in which we collect and store experimental data in-house and what is required to make our data more accessible. Creating such templates during the experimental planning stage would help to outline and standardise the data being collected for each experiment and will lessen the effort required if later modification is needed. Since working on NanoFASE, we have become involved with two further projects, SAbyNA and ASINA, which aim to upload data through the same systems. Since the NanoFASE project experience, we feel prepared to both create the templates and populate them in the correct way. We have decided to create the templates from the start and will incorporate these into the experimental design stage, with the intention of populating templates as data is collected/ received.
Conclusions on the overall achievement
The conclusions of this case demonstrated a clear need for structured and clearly defined data management processes that need to be applied from the onset of a complex, multidisciplinary and multi-partner project. To achieve the optimum result a clear set of steps is required. First and foremost, the entire process needs to commence from day 1 of the project, as the later the start data the harder the process becomes putting a lot of pressure on the data creators. This can lead to loss of either data and/or metadata reducing data quality, interoperability and reusability.
To achieve maximum engagement, strong support from the project’s coordination team is required along with emphasising on the benefits for data creators, along with the inclusion of data shepherd(s) that are able to discuss, understand and communicate the needs and requirements of each user to the rest and how these can be met from the rest of the consortium, where applicable. Data shepherds need to resolve any conflicts in terminology used under very specific contexts and guide towards the use of more generic terms and combination of existing terms to enhance interoperability. This needs to be complemented with a strong hands-on approach and a combination of joint consortium and partner-specific meetings. The former can provide a general overview of the entire process and a common understanding of the needs and requirements within the consortium. The latter focus on the data produced from specific partners, making it easier to understand their responsibilities, the data and metadata needed to be captured and the way to fill in the templates.
In summary, the case provided invaluable insights on the processes needed to manage the data captured in large, multidisciplinary projects to achieve maximum data quality, completeness and FAIRness. This can only be achieved with the strong support of the project leadership and the engagement of the entire consortium by analytically presenting the benefits of the entire process along with their respective responsibilities. Having the data shepherds organising the practical work helped the scientists to better understand benefits of harmonised, interoperable and FAIR data sharing, becoming obvious from the testimony of the data providers, and, in this way, reducing the activation barrier to invest the needed time to prepare metadata and data accordingly. This understanding was created by the data shepherd executing the following key tasks:
- Listing of all types of data and metadata to be produced during the project’s lifetime through detailed communication with individual partners
- Creation of the data workflows taking into account the specific data curation and warehousing solutions selected by the project
- Organising data management workshops presenting the concepts and the templates used at the earliest stage possible
- Workshops with individual groups to be able to take specific details of the produced data into account
- Hands-on session with data collectors for filling the metadata and data into the data templates
- Continuous identification of suboptimal parts of the curation and management process due to missing tools, user-unfriendly templates or user interfaces, and/or approaches very different from the normal working behaviour in the laboratories
- Design of solutions to remove pain points together with the data warehouse providers and software engineers.
However, even with this support, NanoFASE still started the project-wide data collection and management process too late. Data providers from NanoFASE are now helping to change this behaviour in new projects, in which they are involved in, by performing some of the tasks of the data shepherds. This will be further supported by NanoCommons offering data shepherd services but also by testing new approaches to provide tools, which support documentation of the study design and SOP development and provide information on these steps in form of metadata (see new set of demonstration case below), which can be linked to the final datasets pushing aspects of data management already into the first step of the data life cycle and all downstream steps.
References
- Papadiamantis et al., 2020: Papadiamantis, A. G.; Klaessig, F. C.; Exner, T. E.; Hofer, S.; Hofstaetter, N.; Himly, M.; Williams, M. A.; Doganis, P.; Hoover, M. D.; Afantitis, A.; Melagraki, G.; Nolan, T. S.; Rumble, J.; Maier, D.; Lynch, I. Metadata Stewardship in Nanosafety Research: Community-Driven Organisation of Metadata Schemas to Support FAIR Nanoscience Data. Nanomaterials 2020, 10 (10), 2033. https://doi.org/10.3390/nano10102033.

