dmp

Table of contents
Summary
List of abbreviations
Introduction
Data set description
Data sharing
Archiving and preservation
The
1. Data summary
1.1 Purpose of the data collection/generation
1.2 Relation to the objectives of the project
1.3 Types and formats of data
1.3.1 Types of Data
1.3.2 Formats of Data
1.4 Reuse of data
1.5 Origin of the data
1.6 Expected size of the data
1.7 Utility of data and models
2. FAIR data
2.1 Making data findable, including provisions for metadata
2.1.1 Making data discoverable, including provisions for metadata
2.1.2 Identifiers and naming conventions
2.2 Making data openly accessible
2.2.1 Open Data
2.2.2 Free Access
2.3 Making data interoperable
2.3.1 Supported data exchange formats
2.4 Increase data reuse (through clarifying licenses)
3. Allocation of resources
4. Data security
5. Ethical aspects
6. Other
7. Final remarks
8. References
Appendixes
Appendix A: RDM Copyright, License, and Waiver Clearance Form
Appendix B:
Summary
This deliverable presents the data management plan (DMP) for the
community research infrastructure
The initial DMP covered a significant part of the life cycle of research data,
but could not cover its
life after the end of this project. It covered the initial process of
thinking about how research data will be captured and handled during the
research, networking and transnational activities (JRA, NA and TA,
respectively), the follow up process of making it Open and FAIR, and
finally the long-term deposition of the data (and models and other outputs),
to ensure a life after the end of the
Therefore, this deliverable continues to describe suggestions for how the
project partners and TA users can make data available in both Open and
FAIR formats. Note that the
List of abbreviations
APIs - Application Programming Interface
ASTM - American Society for Testing and Materials
CEN - European Standardisation Organisation
ChEBI - Chemical Entities of Biological Interest
CLW - Copyright, License, Waiver
DoA - Description of Action
DMP - Data Management Plan
DOI - Digital Object Identifier
EC - European Commission
ECHA - European Chemicals Agency
EFSA - European Food Safety Authority
EFO - The Experimental Factor Ontology
ENM - Engineered Nanomaterial
EMA - European Medicines Agency
EOSC- EU Open Science Cloud
ERIC - European Research Infrastructure Consortium
EUON - European Union Observatory for Nanomaterials
FAIR - Findable, Accessible, Interoperable, Reusable
GDPR - General Data Protection Regulation
HGNC - Human Genome Nomenclature Committee
IPR - Intellectual Property Rights
ISO - International Standards Organisation
JRA - Joint Research Activities
JRC - Joint Research Center
MoU - Memorandum of Understanding
MPATH - Mouse Pathology Ontology
NA - Networking Activities
NCBI - National Center for Biotechnology Information
NCIT - National Cancer Institute Thesaurus
NEMs - New and Emerging Materials
NM - Nanomaterial
OAE - Ontology of Adverse Events
OBI - Ontology for Biomedical Investigations
OECD - Organisation for Economic Cooperation and Development
ORD - Open Research Data Pilot
PATO - Phenotype And Trait Ontology
QA - Quality Assurance
QC - Quality Control
QSAR - Quantitative Structure-Activity Relationships
R&D - Research and Development
RDM - Research Data Management
RDMO - Research Data Management Online (Plan)
RDS - Research Datastore
SME - Small to Medium Enterprise
SOP - Standard Operating Procedure
TA - Transnational Access
UO - Units of Measurement Ontology
WWTP - Working Party on Manufactured Nanomaterials
Introduction
The European Commission (EC) is running a flexible pilot under Horizon 2020 called the Open Research Data Pilot (ORD Pilot). The ORD pilot aims to improve and maximise access to, and reuse of, research data generated by Horizon 2020 projects and ask projects to think about the totality of their data in all forms, taking into account the need to provide open scientific information, undertake commercialisation activities and Intellectual Property Rights (IPR) management, address data privacy concerns and ensure data security, as well as data management and preservation questions [1]. Open data is data for which everyone has the rights to access, reuse, repurpose, and redistribute. The ORD Pilot aims to make the research data generated by selected Horizon 2020 projects accessible with as few restrictions as possible, while at the same time protecting sensitive data from inappropriate access [2]. Projects starting from January 2017 are by default part of the ORD Pilot, including the Research infrastructures and e-Infrastructures, and as such are required to develop a Data Management Plan (DMP).
To help optimise the potential for future sharing and reuse of data, the
DMP helps the project partners and Transnational Access (TA) Users
to consider any problems or challenges that may be encountered in making their
data Open and FAIR and helps them to identify ways to overcome these. This DMP
is a “living” document outlining how the research data collected or generated
will be handled during and after the
Data set description
This section refers to what kinds of data
- The data and metadata needed to validate results in scientific publications;
- The data and metadata needed to develop and validate the predictive in silico models for nanosafety, via the Joint Research Activities (JRA);
- Data and metadata generated by Users through TA activities; and,
- Other curated and/or raw data and metadata that may be required for validation purposes or with reuse value.
Further, these questions are addressed in order to determine the potential reuse value of the data:
- What is the data about?
- Who created it and why?
- In which forms it is available?
- What (if any) standards were applied in generating the data?
The metadata provided with the datasets answers such questions to enable data to be found and understood, ideally according to the particular standards applied. Finally the metadata, documentation and standards will help in making the data FAIR (Findable, Accessible, Interoperable, and Reusable) [4-6].
Data sharing
According to the ORD Pilot programme, by default as much of the resulting data as possible should be archived as Open Access. Therefore, legitimate reasons for not sharing resulting data should be explained in the DMP. However, data protection or IPR agreements should not be compromised in any way, and data sharing should be done responsibly. Therefore, the DMP describes any ethical or legal issues that can have an impact on data sharing.
Archiving and preservation
To ensure that publicly funded research outputs can have a positive impact on
future research, for policy development, and for societal change it is important
to assure the availability of data for a long period beyond the lifetime of the
project. However, this does not refer only to storage in a research data repository,
but also the need to consider the (re-)usability of the data. One of the main goals
of the research infrastructure being created by the
is that all datasets imported into the
The structure of the
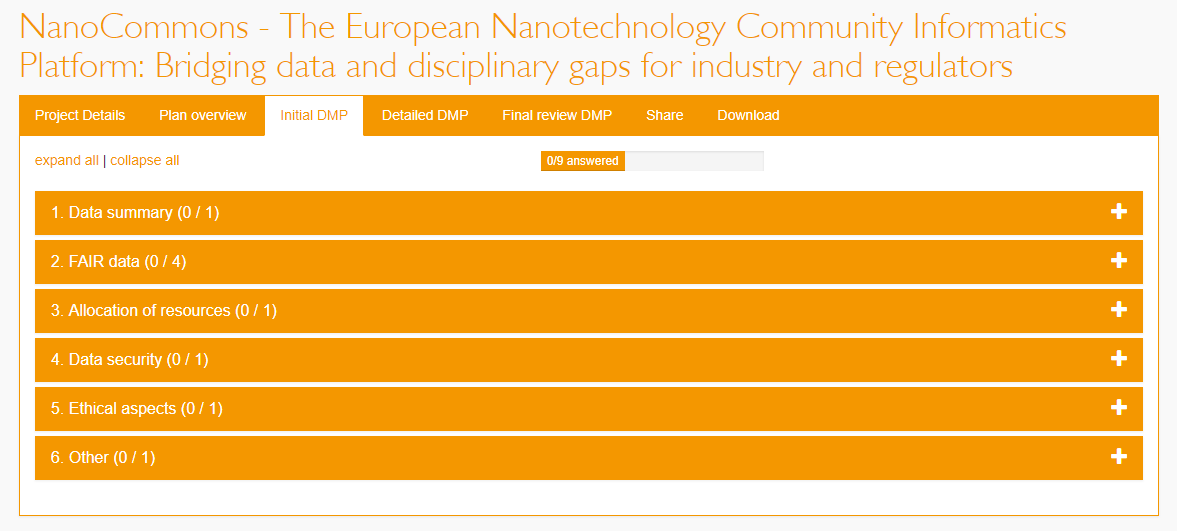
Figure 1. Screenshot of the tool used to create the
The Data Stewardship Wizard is another tool to support data management planning.
Example: Archive and get a free DOI
NanoCommons as well as other EU NanoSafety Cluster project have been archiving openly-licensed datasets, making the released data available for a longer time. For example, Zenodo and Figshare guarantee 20 years of availability of the data. They also give data a Digital Object Identifier (DOI) making data citations easier. Of these two, Zenodo has the advantage for H2020 projects to have integration with the CORDIS database, and research output can be directly linked to the grant.
NanoCommons has been collecting openly, citable data output from the cluster and made this available on a dedicate webpage at nanocommons.github.io/datasets/ (see Figure 2) and embedded Bioschemas annotation, allowing search engines to discover the data sets, including the Google Dataset Search.
 Figure 2. Screenshot of the NanoCommons overview of openly-licensed
data sets produced, collected, or created by EU NanoSafety Cluster projects.
Figure 2. Screenshot of the NanoCommons overview of openly-licensed
data sets produced, collected, or created by EU NanoSafety Cluster projects.
The
The Data Management Plan (DMP) is essential to ensure the capture of
return-on-investment (RoI) for the resources put in by the European Union,
via the EC’s H2020 research framework. An RDM strategy does not describe
the data, but it describes the processes of the management of the data
throughout its entire lifecycle. The
Before we continue, we should define what kind of data this DMP
applies to. Section 1.3 provides more detail of the types of data to be
imported into or generated within
Expectations
The
A specific strategy to provide open access to data will be developed according to the recommendations of the Horizon2020 ORD Pilot. The strategy will include the development of initial and final FAIR DMPs (M6 and M48 respectively), which will outline and support implementation of procedures to ensure full access to data incorporated within
However, a second section of our DoA is essential, and is part of our agreement (contract) with the EC, which describes in more detail how
29.3 Open access to research data
Regarding the digital research data generated in the action (‘data’), the beneficiaries must:
a. deposit [it] in[to] a research data repository and take measures to make it possible for third parties to access, mine, exploit, reproduce and disseminate — free of charge for any user — the following:
i. the data, including associated metadata, needed to validate the results presented in scientific publications as soon as possible;
ii. other data, including associated metadata, as specified and within the deadlines laid down in the ‘data management plan’;
b. provide information — via the repository — about tools and instruments at the disposal of the beneficiaries and necessary for validating the results (and — where possible — provide the tools and instruments themselves).
This does not change the obligation to protect results in Article 27, the confidentiality obligations in Article 36, the security obligations in Article 37 or the obligations to protect personal data in Article 39, all of which still apply. As an exception, the beneficiaries do not have to ensure open access to specific parts of their research data if the achievement of the action’s main objective, as described in Annex 1, would be jeopardised by making those specific parts of the research data openly accessible. In this case, the DMP must contain the reasons for not giving access.
These formal expectation set aside, the project has quite some liberty in setting our own expectations, and these may even be different from one partner to another. All partners have their legal obligations and the
1. Data summary
Summary of the data addressing the following issues:
- State the purpose of the data collection/generation
- Explain the relation to the objectives of the project
- Specify the types and formats of data generated/collected
- Specify if existing data is being reused (if any)
- Specify the origin of the data
- State the expected size of the data (if known)
- Outline the data utility: to whom will it be useful
1.1 Purpose of the data collection/generation
The data collected/generated within the
project aims to implement appropriate standardised experimental procedures and data curation techniques, to promote ontology harmonisation and produce datasets aligned with the highest standards of scientific quality within the project.
Thus, the data collected/generated within
1.2 Relation to the objectives of the project
is driven by the European nanosafety, nanomedicine and emerging materials research and regulatory communities need for an e-infrastructure providing a standardised, reproducible and interoperable way to access all available data, knowledge and analysis and facilitate the application of nanoinformatics and modelling tools that have been adapted and verified as suitable for application to nanomaterials.
More than 10 years of nanosafety research has delivered tangible insights into the key science and policy required for the development of safe nano-enabled products. However, this knowledge has yet to be systematised, or made “FAIR”, in a manner that allows:
- modellers to develop predictive frameworks and assess their domains of applicability,
- industry to utilise the data, models and tools for safe-by-design strategies or as supporting evidence for use in regulatory dossiers,
- regulators to compare one form to another or make estimations of data requirements for New and Emerging Materials (NEMs) based on shared properties with NMs, or
- educators to utilise in teaching toxicology, ecotoxicology, environmental fate and modelling of the behaviour of ENMs.
To address this gap,
will facilitate:
- the efficient collection, integration, curation and maintenance of existing data and methods along with development and optimisation of the tools and user interfaces to interrogate them (JRA),
- the provision of access to the data, methods and tools collected or produced under the project, along with expert guidance in their use and in experimental design and workflows to harmonise data quality into the future (TA), and
- community building including bridging disciplinary gaps (e.g. toxicology and ecotoxicology, experimental and modelling), promoting best practice in data quality (e.g. Quality Assurance (QA) audits, Independent Experimental Data audits), and development of User case studies demonstrating the capability of the
infrastructure to address real stakeholder challenges in partnership with industry & regulators (NA).
is designed to integrate the Knowledge infrastructure for risk assessment of novel and emerging materials on a European and international scale, and provide (remote) access to data, data mining, modelling and risk assessment tools to all European researchers, from academia and industry, as well as regulators, ensuring their optimal use and joint development.
1.3 Types and formats of data
1.3.1 Types of Data
An important distinction should be made here between various forms of data to be generated and utilized within
- Raw or experimental data
- Derived (processed or computed) data
- Data associated with formal publications (literature curated data) (This is typically summarised data, from which the original data cannot really be reconstructed).
- Interactive data, whereby the data is put in the context of other data. For example, integrated with other data sets, etc.
The first type of data is basically what comes directly from the instruments utilised in the experimental assays or the computational calculations. Ideally this follows the design of a pre-registration, in which the design of the experiment is published before the experiment is performed (see e.g. the Center of Open Science Pre-registration Challenge: cos.io/prereg/). The second type is data derived from the raw data, and aimed at making comparisons between experimental conditions, drawing conclusions, and other kinds of use and reuse of the data. This data may be stored as spreadsheets, but also in many other formats.
The third and fourth types of data are more about presentation of the data: the third kind is the presentation in formal journal applications and, for example, presented in tables as PDFs, spreadsheets, or data files. The fourth kind is particularly interesting and important to our project: data must be interactively available to enable reuse. For example, our modelling tasks depend on data to be available in a FAIR way (see below).
The Open Science expectations around data basically apply to all four kinds of data. However, it is clear that different solutions are needed for the different kinds. This is one reason why writing a clear DMP is non-trivial. The use of electronic notebooks (for both experimental and computational workflows) will ensure that data of types 1 and 2 above will be collected in a harmonised, ontology-linked, and database-compatible manner from the outset, thereby integrating data management with data generation, rather than data management being an add-on activity after associated with, for example, publication requirements for datasets to be deposited in appropriate databases. Free and commercial tools for keeping an electronic notebook are both available.
Data Life Cycle
The Data Life Cycle includes the entire process through which data is generated, acquired, analysed, manipulated and made available through publications and/or data repositories along with the metadata produced during the entire process. The fourth kind of data presented in the previous paragraph can also be seen as an overview of the life cycle of data. However, it should be noted that the availability of data does not constitute the start of the cycle: instead, the design of an experiment is a more appropriate start - see Appendix B for further details of the data life cycle as utilised in
1.3.2 Formats of Data
The
1. ISA (-tab or -json)
a. The advantage of the use of the ISA type templates is the flexibility they demonstrate with respect to creation and design for any type of experiments and the addition / subtraction of columns to fit all experimental needs (methods, descriptors etc.).
b. Disadvantages of the ISA- templates include the lack of description of the file formats for both data and metadata. They are also not necessarily linked with specific ontologies and thus the naming of the columns is not regulated and protocolled. At the same time, only the file names, and not the file types, are generally available. The latter disadvantage could be potentially overcome with the use of an interoperability layer within the data management infrastructure, which will describe the dataset using a specific ontology and taxonomy.
c. The ISA-(TAB/JSON)-nano addon is currently not as developed [8], but is the started norm in the nano-field, although is rarely applied to the letter. ISA- (TAB/JSON)-nano is being promoted by the US National Cancer Institute and the eNanoMapper data portal.
2. NIKC
a. The advantages of the NIKC template, which is an extension of the ISA-tab nano, are its dynamic nature and design flexibility, being able to facilitate all aspects of nanosafety research (e.g. nanomaterials characterization including transformations and ageing, exposure, environmental as well as human hazard and risk assessment, regulatory), while accepting the attachment of images. The currently developed NIKC/
b. The disadvantages of the NIKC templates are that are complicated and time consuming to design and build. They also span at several different tabs that require the logging of a large amount of information. As a result, they need extended training from experienced curators that are also tasked to QC the produced templates. Some of the issues could be circumvented with the development and use of an automated system of template creation, based on a questionnaire system and having a predefined set of nanomaterials characterisation template. Additionally, as more and more study templates are developed, reuse will be possible with potentially minor tweaks.
3. ToxML
a. The advantage of the ToxML data format is the robust design of the files, which also simplifies interoperability. The latter, in certain cases, can also be considered as a disadvantage.
b. The drawbacks for the use of ToxML is the difficulty to implement changes, which have to proposed and then reviewed and accepted by the supervisory board making the process extremely time consuming. As a result, the
In all cases, the usage of a common ontology for data description and curation can guarantee that both the data and produced metadata can be understood by both users and machines and can be automatically transferred between services.
1.4 Reuse of data
aims to use the data collected/generated through the project, as well as high quality curated peer-reviewed published data. In terms of tools, the project will identify and use already existing tools (modelling, Quantitative Structure-Activity Relationships (QSARs), Omics etc.) and will work to improve them further using the data collected/generated through the project. The
For data templates, SOPs and decision trees, we will use, as much as possible, existing data, software tools, open and readily available to all partners. We will aim to produce reusable and extensible tools, and to make use of existing nanomaterials safety and characterisation data, where possible.
SOPs developed within NanoMILE and NanoFASE for characterisation of NMs in biological and environmental matrices are being reused where possible, and any changes documented as part of the metadata and domains of applicability sections of the SOPs. For example, in the development of SciNote online notebooks, we are utilising existing lab data and lab protocols.
Table 1 below summarises the datasets that are potentially being reused within the context of
Table 1: Existing data generated through other NanoSafety Cluster (NSC) projects and other appropriate projects will be used by the
| Project | Description of Data | Purpose in |
|---|---|---|
| NanoMILE | Characterisation data and calculated descriptors for up to >75 different NMs via the NanoMILE NMs library. Data identifying ENM property factors important for hazard, and representative ENMs that range in those properties. | To provide baseline characterisation data for a wide panel of NMs, that will be used to parameterise the domain of applicability of the various computational models, as well as for benchmarking of the newly developed models and tools. |
| NanoFASE | Fate process relevant data (e.g. separation between sludge and effluent in WWTPs) and characterisation of the transformations of NMs in the different environmental compartments. Data on microbial degradation of commonly applied coating materials. | Considerable method development underway, specifically for functional assays to characterise the transformed states of nanomaterials in different environmental compartments. This data will be utilised to demonstrate the validity and utility of the NIKC instances approach, and how the concept expands dramatically the scale of the data available, but also the predictions that can be made utilising the data. |
| NanoPolyTox, GUIDEnano, SUN | Release methods and data for NMs enabled product types especially of worked case studies. | Release data will be utilised to support the |
| NIKC | Characterisation data of nanomaterials through their entire lifecycle in conjunction with characterisation of the surrounding environment. Anticipated use scenarios, matrix, concentration in products, system characteristics (environmental, biological, laboratory, etc.). Exposure and hazard measurements, calculations, and estimates and metadata associated with each of these, including bibliometrics, protocols, equipment, temporal and spatial descriptors, etc. | The currently curated hazard and exposure data will be utilised to support the |
| NANoREG | Characterisation data for the JRC repository test NMs, and the templates for data collection for selected characterisation end-points. | The characterisation data are deposited in the eNanoMapper database, and an MoU will be sought to access the characterisation data as part of the Model benchmarking and Domain of applicability assessment. |
| eNanoMapper | Ontology terms, database structure for nanosafety. | |
| Projects funded under NMBP-14-2018 (nanoinformatics call) | The projects funded under this call will be specifically developing nanoinformatics models, and will also require a knowledge warehouse, APIs, and large scale datasets, so we will provide access to |
By facilitating access to and reuse of datasets that it has compiled, curated, integrated and aligned, |
1.5 Origin of the data
The data sources and offered tools through the project will take into account the original licences for the versions integrated into the
- Data, models and tools developed and owned by
Partners will be assigned with a Creative Commons licence, allowing their full and free reuse, modification, and redistribution, where applicable, as long as their origin is cited appropriately as far as possible. When only non-commercial reuse is allowed, then a specific, well-justified case needs to be submitted and approved by the coordinator under the terms of the Consortium Agreement; - Open Source data, tools and models, used the license mentioned by the owners;
- Data from third parties, and not yet available in existing open databases used under the conditions specified by the data owner and included in a formal agreement.
1.6 Expected size of the data
The data generated/collected through the project’s open calls, and produced through partner and user collaboration, will be in the region of 10s to 100s Terabytes and will consist of raw, analytical and metadata, and the databases to support the project’s actions. Data access, during and beyond the project’s life cycle, will be facilitated through processes that will ensure that all data will be stored in the project’s centrally managed datastore, i.e. the
In the case when data or tools available online from external sources will be required accessibility will be achieved through its original source, along with the implementation of a harmonisation system layer to ensure data and tool interoperability with that of the
The data to be made directly publicly accessible is the description of the models and tools themselves, along with the SOPs, analysis of uncertainty, domain of applicability and benchmarking against other relevant methods / models / tools. Guidance on the use of the different models and approaches, appropriate input data and how to generate the appropriate input data, and data capture templates will also be developed and integrated. The size of this data may be comparatively limited and thus easier to make interoperable and reusable.
A key aspect of the
1.7 Utility of data and models
FAIR data are at the core of
The offered services can be used in a way to maximise the potential of subsequent use in the standardisation of novel approaches, via OECD, CEN, ISO, safer-by-design etc. further supporting the utilisation by academics, industry and regulators. The harmonised interoperability between academia and industry can provide a wide amount of data allowing innovative R&D and computational activities and lowering the barriers of real innovation resulting in new safer and secure products, processes and services. The close cooperation with the 3rd pillar of nanosafety, regulators, is also key to promoting the wider acceptance of the produced workflows, tools and services.
Potential beneficiaries of the data, tools and the
- Academics, at all levels, working in all fields of nanosafety research and the wider toxicity community. The services offered can help them uncover underlying research patterns and reach new scientific conclusions.
- Regulatory agencies (e.g. ECHA, EMA, EFSA) and policy makers.
- SMEs that do not have the resources or the knowledge to develop and use in-house tools for safer-by-design approaches and risk assessment requirements.
- Industry and the R&D community, which can use the offered services to address the ‘3Rs’ principles and let them design novel and safer experimental approaches.
- Consumers, through the interoperability of all of the above that will offer them new safer products containing nanomaterials.
2. FAIR data
The FAIR principles refer to a number of features that data, software, etc. should have to maximize their value and societal impact [11]. They are grouped into four categories, as given before. Each of the four aspects of the principles will see a different way it is implemented for that kind of data. For example, in some cases, raw data may not be findable to people outside
How these principles are implemented, how they are used, is totally up to the user. They have been defined quite broadly so that apply to different kinds of scientific output. This has led to confusion about how to make your data FAIR. In fact, it is not a black-and-white situation, but there are many shades of grey. The point is that data should be as FAIR as possible. This in turn suggests there is a scale of FAIR-ness, and metrics have been proposed [4].
2.1 Making data findable, including provisions for metadata
- Outline the discoverability of data (metadata provision)
- Outline the identifiability of data and refer to standard identification mechanism. Do you make use of persistent and unique identifiers such as Digital Object Identifiers?
- Outline naming conventions used
- Outline the approach towards search keyword
- Outline the approach for clear versioning
- Specify standards for metadata creation (if any). If there are no standards in your discipline describe what metadata will be created and how
2.1.1 Making data discoverable, including provisions for metadata
This principle prescribes that the output must be findable. That is, effort must be made to make sure people can find the data. To be findable, the principles specify:
- F1. (meta)data are assigned a globally unique and persistent identifier
- F2. data are described with rich metadata (defined by R1 below, in section 2.4)
- F3. metadata clearly and explicitly include the identifier of the data it describes
- F4. (meta)data are registered or indexed in a searchable resource
To meet these principles the project aims to use data repositories to store the data generated/collected by the Project and the tools developed from the Consortium. This will allow the datasets and tools to be easier to find and access. The repositories will also include an appropriate licensing system to allow a layered access to the available data based on the accessibility decided by the data and/or tools owners. Both datasets and tools will be complemented with the appropriate metadata (and source code in the case of software tools whenever possible) and unique repository and/or DOI identifiers to allow users to easily query and reference. The Project also aims to establish collaborations with existing and under development data repositories and link them together through a joint query system. To achieve this, necessary harmonisation between the repository outputs needs to take place to allow the queried results to be returned in a consistent manner, which will also promote interoperability.
As a result,
- accessing a query system with a filtering functionality;
- to identify the specific dataset’s/tool’s licensing and access information, desired data fields to be searched (e.g. ENM characterisation parameters, toxicity results, tools source code);
- a harmonised data exchange file format; and,
- full access to the necessary metadata, which will describe the experimental setup, the analytical tools used and a summary of the research outputs.
To achieve that harmonised output,
2.1.2 Identifiers and naming conventions
Of particular importance are the use of identifiers here. As many aspects of the experiments should be linked to identifiers, including but not limited to: ENMs, cell lines, bioassays, species, genes, proteins, and metabolites. Identifiers should come from internationally recognized databases or resources whenever possible. For example, gene identifiers should be ideally from Ensembl or NCBI Gene, or from the HGNC for human genes. For proteins UniProt would be a good resource. For nanomaterials, the proper JRC identifiers (JRCNMxxx) must be used (the NM-xxx only apply for very old batches) [12]. If nanomaterials do not have globally unique identifier, the
2.1.2.1 Disseminate testing material identifiers
Similar to the idea of pre-registration of clinical trials, each project should disseminate the identifiers they have chosen to represent the nanomaterials selected to be tested or used in that project. These identifiers must be publicly announced as soon as a material is selected, widely distributed within the project, disseminated to other projects, and consistently used in all reporting, experimental protocols, results, metadata, etc.
To support this, the European Registry of Materials was started where anonymous identifiers can be registered, called an ERM identifier [21]. These are then reserved and can be used in all (internal and external) documentation (experimental designs, spreadsheets, reports, deliverables, articles, etc), e.g. as Compact Identifiers (see Section 2.1.2.2). Registration of an new identifiers requires only a minimal amount of information: a name for the material, e.g. “NanoCommons Material X”. Optional information includes: unique chemical composition, batch and/or lot number, an ontological classification, a webpage, a provider, contact, or project name.
2.1.2.2 Use IRIs or Compact Identifiers in dissemination
To ensure clear and explicit semantics, when using identifiers in dissemination, either full identifiers should be used, such as Internationalized Resource Identifiers (IRIs) or, alternatively, Compact Identifiers [20]. The ERM identifier identifier can be used to identify nanomaterials in datasets as IRI but also as Compact Identifier or CURIE in written material. For example, the caLIBRATE project has registered the erm:ERM00000074 identifier, using the ‘erm’ prefix in this CURIE/Compact Identifier notation, but has the full IRI https://nanocommons.github.io/identifiers/registry#ERM00000074.
2.1.3 The InChI for Nano
Another identifier was explored, based on the IUPAC International Chemical Identifier (InChI) [22]. Once fully developed, the nano InChI can be used as a global, unique identifier. Currently, this is not advised yet. An InChI Trust working group has been set up for the further development (see inchi-trust.org/Nanomaterials/).
2.2 Making data openly accessible
- Specify which data will be made openly available? If some data is kept closed provide rationale for doing so
- Specify how the data will be made available
- Specify what methods or software tools are needed to access the data? Is documentation about the software needed to access the data included? Is it possible to include the relevant software (e.g. in open source code)?
- Specify where the data and associated metadata, documentation and code are deposited
- Specify how access will be provided in case there are any restrictions
This principle prescribes that data must be accompanied with metadata that explains how people get access to the data. This does not imply they always get access (generally they do), but if they do, how. To be accessible, the principles specify:
- A1. (meta)data are retrievable by their identifier using a standardized communications protocol
- A1.1 the protocol is open, free, and universally implementable
- A1.2 the protocol allows for an authentication and authorization procedure, where necessary
- A2. metadata are accessible, even when the data are no longer available.
2.2.1 Open Data
The aforementioned section from our DoA briefly outlines the git of the H2020 Open Data pilot: we give access to third parties to our data. In fact, the section is quite close to the core rights involved in Open Science, the rights to use/reuse, modify, and reshare knowledge. At the same time it does set some reasonable limits: for example, where there are good reasons to not make data available (e.g. because of privacy aspect of human data), this is acceptable but must be explicitly reasoned for, and this must be documented. However, this guidance leaves plenty of room for us to implement our approach, and this leaves plenty of options and decision to be made.
2.2.2 Free Access
One controversial aspect of the current implementation of Open Access is to provide free access, free as in without cost to the user. Of course, hosting data is not without cost, but
To place the
In the long run,
But generally it is accepted that hosting raw can be covered in kind by existing solutions:
- Institutional repositories (e.g. UoB BEAR Hosting Services))
- European repositories (e.g. ZENODO, part of OpenAIRE)
- Commercial repositories (e.g. Figshare or Mendeley Data)
Guidance on selecting repositories can be found in Science Europe’s Practical Guide to the International Alignment of Research Data Management.
2.3 Making data interoperable
- Assess the interoperability of your data. Specify what data and metadata vocabularies, standards or methodologies you will follow to facilitate interoperability
- Specify whether you will be using standard vocabulary for all data types present in your data set, to allow inter-disciplinary interoperability? If not, will you provide mapping to more commonly used ontologies?
This principles prescribes that data must be sufficiently annotated such that people can understand what the data means. This means a clear data format must be used, values must have units, columns must have clear annotation what was measured, etc. This is also where ontologies (vocabularies) typically come in. To be interoperably, the principles specify:
- I1. (meta)data use a formal, accessible, shared, and broadly applicable language for knowledge representation
- I2. (meta)data use vocabularies that follow FAIR principles
- I3. (meta)data include qualified references to other (meta)data
The
Another important aspect of data harmonisation is the use of a vocabulary (i.e. ontology) that will employ common agreed definitions for the terms used by all aspects of nanosafety research and will allow both qualitative and quantitative data combination and reusability. As a result, the mid to long-term plan of
Data and tools storage and accessibility is also going to be addressed through the use of multiple backups and mirrors that will be accessible through the
In any case,
2.3.1 Supported data exchange formats
It is recommended to discuss with
Most database come with other supported formats and exploring the formats in which data will be shared is an essential part of a DMP. For data hosted with eNanoMapper technology, various formats are supported. These include a custom Resource Description Framework format (for which a tutorial is being developed), and various spreadsheet templates (IOM templates [14] and JRC Templates [15]). The latter two take advantage of a spreadsheet annotation tool developed in eNanoMapper, called nmdataparser. This approach has been used to make NANoREG data available to the NanoSafety Cluster community.
For long term data storage the ISA formats may be of interest. Options are then the original ISA-Tab format, including the ISA-Tab-Nano extension, and the newer JavaScript Object Notation (JSON)-based ISA formats, for which also a nanomaterial extension has been developed [16].
Of course, for the many omics data types, domain specific formats are recommended, along with deposition of Open Data in domain repositories with those formats.
2.4 Increase data reuse (through clarifying licenses)
- Specify how the data will be licensed to permit the widest reuse possible
- Specify when the data will be made available for reuse. If applicable, specify why and for what period a data embargo is needed
- Specify whether the data produced and/or used in the project is usable by third parties, in particular after the end of the project? If the reuse of some data is restricted, explain why
- Describe data quality assurance processes
- Specify the length of time for which the data will remain reusable
This principle basically outlines that community practices for data sharing and data science should be followed. To be reusable, the principle specify:
- R1. meta(data) are richly described with a plurality of accurate and relevant attributes
- R1.1. (meta)data are released with a clear and accessible data usage license
- R1.2. (meta)data are associated with detailed provenance
- R1.3. (meta)data meet domain-relevant community standards
Data reusability is in the core of
envisages to also link to its database high quality data from other publicly available sources spanning the full spectrum of nanosafety research. To achieve that the Project needs to harmonise the data as much as possible, develop an appropriate query system and make it available to third parties under the produced licensing system. Such an action means that the data and tools under offer needs to be systematically checked using strict QA processes.
QA processes for data generated/collected by
A similar approach needs to be implemented for pre-existing datasets to be imported into the
3. Allocation of resources
Explain the allocation of resources, addressing the following issues:
- Estimate the costs for making your data FAIR. Describe how you intend to cover these costs
- Clearly identify responsibilities for data management in your project
- Describe costs and potential value of long term preservation
The Project already runs a survey that tries to monitor the awareness of the nanosafety community on FAIR data and aims to promote it through targeted workshops.
It is the intention to minimise the costs by using free-to-use data repositories and dissemination facilities to achieve GREEN open access, for example OpenAIRE, the UoB and NERC Open Research Archives etc. Some partners have foreseen and budgeted funds to get a few high impact papers into GOLD open access journals where the increased impact from this would warrant the expense.
The responsibility for collation and entry into the
The responsibility and budget for developing and managing the web based structure (including aiding partners with data entry and uploading) of the
Transnational Access (TA) to Knowledge Management and computational Tools
Within
Knowledge Base and Data Mining
Processing and Analysis services
Predictive nanoToxicology
Risk assessment visualisation and reporting
Nanoinformatics workflows for nanosafety experiments / best practice workflows.
All TA will be offered via centralised calls for User Access run on a 6-monthly basis. Calls for access will include micro-projects (up to 1 month Access), mini-projects (up to 6 months of access) and workflow Access to include site-visits to expert labs and support in establishment of nanoinformatics workflows to underpin excellence in experimental design. A prerequisite for TA projects related to TA modality will be that data utilised will be made open access and FAIR via
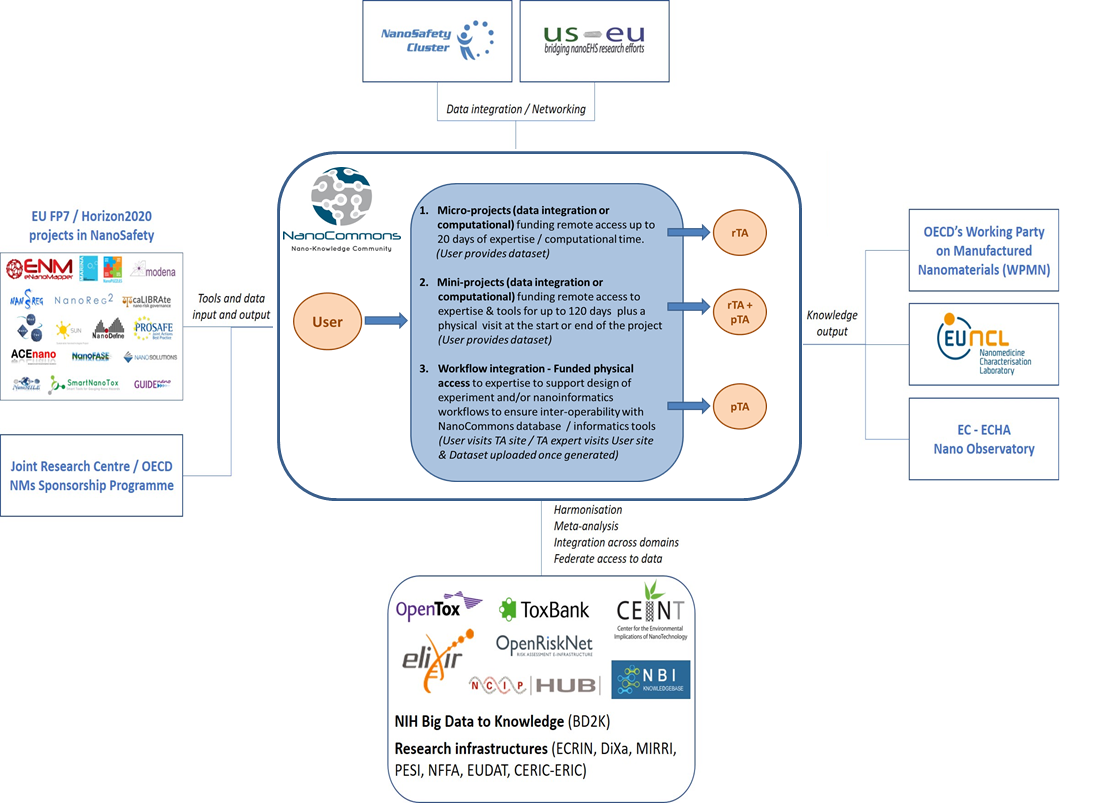
Figure 2. Flow of data in the
4. Data security
- Address data recovery as well as secure storage and transfer of sensitive data
The
- Responsible and fully secured management processes for personal data. These will include the anonymisation, encryption, logging of data usage as well as data deletion following usage. Implementation has already begun with the
website having all necessary security protocols and tools (anti-hacking and malware plugins) added to prevent any malicious attacks. - The management team and the
Consortium will ensure that all partners and users will follow strict ethical guidelines covering all aspects of the project’s infrastructure. A privacy-by-design approach will be followed and controlled by an independent Data Protection Officer. - The presence in the consortium of established software and tools development partners (e.g. Biomax, DC) will ensure data protection with the use of their state of the art firewall capabilities that can also protect the entirety of the project’s virtual environment.
Data sharing and transfer among persons or partners will be, where appropriate, third party secure file transfer facilities, such as the
5. Ethical aspects
- To be covered in the context of the ethics review, ethics section of DoA and ethics deliverables. Include references and related technical aspects if not covered by the former
The ethical aspects of the project will cover all aspects of the
No ethical issues arise from the project overall, or the data management aspects, since the focus of the project is on making existing datasets accessible rather than generation of new data. However, a strong effort will be made to ensure that ethical information regarding the datasets integrated into
Thus, we do not anticipate any ethical or legal issues relating to the datasets generated as part of ACEnano project that would impact on data sharing. No personal data will be collected and so informed consent for data sharing and long term preservation of such datasets is not required.
6. Other
- Refer to other national/funder/sectorial/departmental procedures for data management that you are using (if any)
As part of University of Birmingham’s commitment to ensuring FAIR and Open data, all research active staff (Postdoctoral fellows, PhD students) are expected to prepare DMPs for their own data, as per the University’s Research Data Management Policy. The UoB data management policy defines research data as “the evidence that underpins the answer to the research question, and can be used to validate findings regardless of its form.” Thus, data covers quantitative and qualitative statements, raw data from measurements and derived data – either cleaned or extracted from a researcher’s primary dataset or derived from an existing source.
A detailed set of guidance on preparation of DMPs available via the UoB DMP site: intranet.birmingham.ac.uk/as/libraryservices/library/research/rdm/Data-management-plans.aspx
will utilise the following aspects of the UoB data storage services:
- Research Data Store (RDS): The RDS is a central storage service for ‘active’ research data. It is highly resilient and is hosted in two data centres on campus. Space on the RDS is allocated to projects and managed accordingly. Up to 3TB of storage will be allocated by default to the Project though additional capacity may be purchased.
- BEAR DataShare: BEAR DataShare is a file synchronisation and sharing service provided by IT Services. The service allows users to securely save and sync files with colleagues and partners anywhere in the world, from any device. It provides 25GB storage capacity per user.
7. Final remarks
A living DMP
The core of the RDM will be fixed: where our DoA or law outlines specific elements, these elements will be core part of the RDM and be pretty static. That said, over four years, the laws will changes, which will affect our plan. Second, to achieve maximal impact of our project, we have to be able to aptly respond to changes in the field.
Important Laws
European law changes all the time, and while not too fast,
- Copyright laws
- Privacy laws
- GDPR law
Both affect our work. Organizations like the ELIXIR, Electronic Frontier Foundation, and Creative Commons are very knowledgeable in the field, and so do several members of the European Parliament, in particular Julia Reda. IT journals also provide information, like the German C’t with Welche Änderungen die neue EU-Datenschutz-Regulierung in Deutschland bringen wird.
RDM tools
There are many tools around that support and/or provide guidance, including the following services:
- DMPOnline (used in this report)
- Data Stewardship Wizard (ELIXIR-NL)
- RDMO: Research Data Management Organiser
- ReDBox Research Data Management Plan (RDMP) tool
RDM courses and guidance
- H2020 Guidelines on FAIR Data Management in Horizon 2020
- “Data management made simple” [17] with Twelve tips for writing a data-management plan
- ELIXIR Webinar: Requirements in data protection law and the upcoming General Data Protection Regulation (GDPR) implementation
- DataONE Webinar: Data management plans 2.0: Helping you manage your data
- SpringerNature’s Research Data Support
- ORCID and Data Privacy in Germany
- “Support Your Data: A Research Data Management Guide for Researchers” [18]
- Data Protection Officer (DPO) Certification course by Maastricht University
- 12 Things to Know About the GDPR and Data Security
- Complete guide to GDPR compliance by GDPR.eu, a H2020 funded project
- OpenAIRE: a pillar for Open Science in the EU: a Horizon 2020 GRACIOUS Webinar
- Ten simple rules for machine-actionable data management plans (preprint) by OpenAIRE
- The (GA4GH) Data Use Ontology (DUO)
- Science Europe: Practical Guide to the International Alignment of Research Data Management
- “Data management” in ELIXIR TeSS
A recent paper looked into the effect of RDM in research, which is worth reading too [19]. They report three “key takeaways: (1) Most PIs practice internal data management in order to prevent data loss, to facilitate sharing within the research team, and to seamlessly continue their research during personnel turnover; (2) PIs still have room to grow in understanding specialized concepts such as metadata and policies for use and reuse; (3) PIs may need guidance on practices that facilitate FAIR data, such as using metadata standards, assigning licenses to their data, and publishing in data repositories.” These provide clear guidance of mistakes not to make yourself.
Our DMP
The previous sections already outlined the context of our DMP. The vision of
- Acquire knowledge about DMPs
- Develop their DMP
- Describe the life cycle of your data
- Develop where data is stored locally
- Develop when, how, where data is disseminated
- Discuss their the DMP within
to align processes and outputs.
Planning the data life cycle
Before writing an implementation of a DMP for specific research, it is good to identify the context, particularly when the data life cycle starts and ends for that research project. For example, the data life cycle starts when the experiment is designed: this design determines the amount of data that will be generated, with what experiments, and put requirements on what your electronic lab notebook needs to be able to record (e.g. photos of gels or other scanned material?).
Equally important is to know early on how the data will need to be analysed or integrated with other data sources, and what happens with the data after you are done with it. The end of life of data is often not before long after the project ends. Top class data will be important for the next 50 years.
Planning the research data management
The DMP presented in this work helps you with planning your research data management. The effort needed for the planning should not be underestimated; it does not need to be very long, but if taken seriously, it allows you to plan everything more efficiently, saving you time in the long run.
One area where time can be saved, is making data FAIR. Particularly making interoperable benefits from planning. For example, ontology annotation of data is easier when the details are fresh in your mind, e.g. about the standard operating procedures. In fact, identifying with identifiers or ontology terms which cell lines, buffers, etc, to use, can best be done when the experiment is designed.
8. References
- Directorate-General for Research & Innovation, E.C., Guidelines on FAIR Data Management in Horizon 2020 v3.0. http://ec.europa.eu/research/participants/data/ref/h2020/grants_manual/hi/oa_pilot/h2020-hi-oa-data-mgt_en.pdf , 26 July 2016.
- OpenAire. OpenAire. 2018 20 July 2018]; Available from: https://www.openaire.eu/.
- OpenAire. How to create a DMP Plan. 20 July 2018]; Available from: https://www.openaire.eu/opendatapilot-dmp
- Wilkinson, M.D., et al., A design framework and exemplar metrics for FAIRness. Scientific Data, 2018. 5: p. 180118.
- Carbon, S., et al., A Measure of Open Data: A Metric and Analysis of Reusable Data Practices in Biomedical Data Resources. bioRxiv, 2018.
- Corpas, M., et al., A FAIR guide for data providers to maximise sharing of human genomic data. PLOS Computational Biology, 2018. 14(3): p. e1005873.
- DMPOnline. Data Management Plan Online Preparation Tool. 2018 May 2018]; Available from: https://dmponline.dcc.ac.uk/
- Thomas, D.G., et al., ISA-TAB-Nano: A Specification for Sharing Nanomaterial Research Data in Spreadsheet-based Format. BMC Biotechnology, 2013. 13(1): p. 2.
- Langevin, D., et al., Inter-laboratory comparison of nanoparticle size measurements using dynamic light scattering and differential centrifugal sedimentation. NanoImpact, 2018. 10: p. 97-107.
- CreativeCommons. Licensing types. Share your work: Licensing types 2018 [cited 2018 20 July 2018]; Available from: https://creativecommons.org/share-your-work/licensing-types-examples/
- Wilkinson, M.D., et al., The FAIR Guiding Principles for scientific data management and stewardship. Scientific Data, 2016. 3: p. 160018.
- Joint Research Centre, E.C. JRC NANOMATERIALS REPOSITORY: List of Representative Nanomaterials. June 2016 20 July 2018]; Available from: https://ec.europa.eu/jrc/sites/jrcsh/files/JRC%20Nanomaterials%20Repository-List%20of%20Representative%20Nanomaterials-201606.pdf
- Hastings, J., et al., eNanoMapper: harnessing ontologies to enable data integration for nanomaterial risk assessment. Journal of Biomedical Semantics, 2015. 6(1): p. 10.
- Jeliazkova, N., et al., Deliverable Report D3.4: ISA-Tab templates for selected set of common bioassays, in eNanoMapper, B. Hardy, Editor. November 2016.
- Totaro, S., H. Crutzen, and J. Riego Sintes, Data logging templates for the environmental, health and safety assessment of nanomaterials. EU Science Hub, 2017.
- eNanoMapper. Investigation-Study-Assay (ISA): New material schema for ISA-JSON. Nanomaterial characterisation and bioassays data entry templates 20 July 2018]; Available from: http://ambit.sourceforge.net/enanomapper/templates/isa.html
- Schiermeier, Q., Data management made simple. Nature, 2018. 555(7696): p. 403.
- Borghi, J.A., et al., Support Your Data: A Research Data Management Guide for Researchers. Research Ideas and Outcomes, 2018. 4.
- Mannheimer, S. Toward a Better Data Management Plan: The Impact of DMPs on Grant Funded Research Practices. Journal of eScience Librarianship. 2018. 7(3).
- Wimalaratne, S.M., et al., Uniform resolution of compact identifiers for biomedical data. Scientific Data, 2018. 5:180029.
- Van Rijn, J., et al., European Registry of Materials: global, unique identifiers for (undisclosed) nanomaterials. Journal of Cheminformatics, 2022. 14:57.
- Lynch, I., et al., Can an InChI for Nano Address the Need for a Simplified Representation of Complex Nanomaterials across Experimental and Nanoinformatics Studies? Nanomateirals, 2020. 10(12):2493.
Appendixes
Appendix A: RDM Copyright, License, and Waiver Clearance Form
| Who are the copyright owners (names + email addresses)? | |
| Under what conditions will the data be available to the consortium? | |
| Under what conditions will the data be available to the rest of the world? |
The
infrastructure via the project’s TA’s. In such cases
If a CC license is or was used, then it suffices to list the name of the license. In
that case, the answer to the second and third question may, in fact, be the same. In
any other case, data providers will have to agree to the
servers, so that it can facilitate experimental workflows and is readily
available for use by the
Appendix B:
The
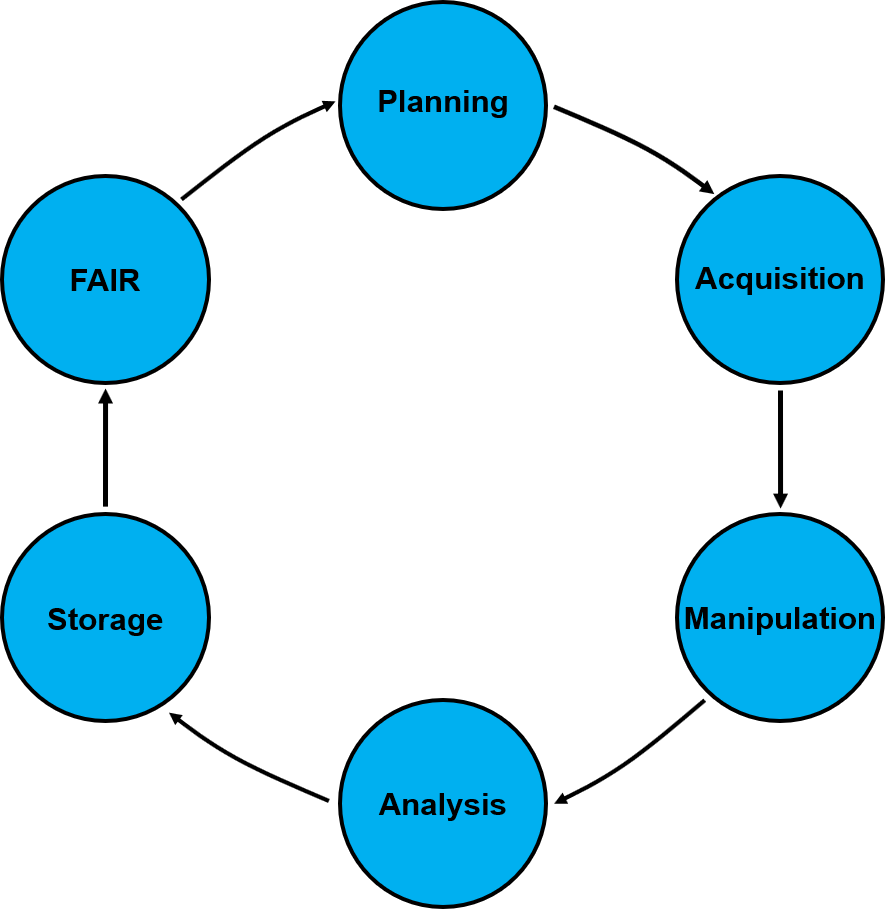
</br>
Figure A1. The complete data lifecycle.
Step 1: Experimental planning
The first step of the data lifecycle is the experimental planning, during which the identification of research endpoints takes place, the number of samples and time points, and the necessary workflow and experimental assays and/or modelling tools are identified. During this step the whole experimental workflow is designed, the detailed experimental and/or analytical protocols identified, what is recorded and in what detail, and the appropriate data curation templates created. This process will take place irrespective of the nature of the needed work (experimental and/or theoretical), as it will customise the workflow to the specific needs of individual Users.
The specified workflow will be implemented into the SciNote online lab-book (Figure A2, scinote.net), which will include all the separate steps of the experimental and/or theoretical workflow. SciNote was the prefered online lab-book due to a combination of user friendliness, dynamic and versatile nature and significant capabilities, although other options are also commercially available. SciNote offers the capability of implementation of a desired data master curation template for experimental data to be stored and extracted, while providing also the opportunity for distinct smaller scale templates, which can be protocol specific. It also offers the capability of analytical protocols implementation, metadata creation and reporting. It also allows the design and assignment of experimental workflows at various levels ranging from an individual researcher to a consortium wide scale, while being readily available both through local (LAN) and wide (WAN) networks based on specific needs.
Each workflow step will be linked to the necessary experimental and/or
computational detailed protocols (Figure A3), the results page (Figure A4)
and the master data curation template (Figure A5), which will be possible
to automatically extract and upload to the desired data repository. The
complete experimental or computational stepwise workflow will be accessible
by the TA Users and the
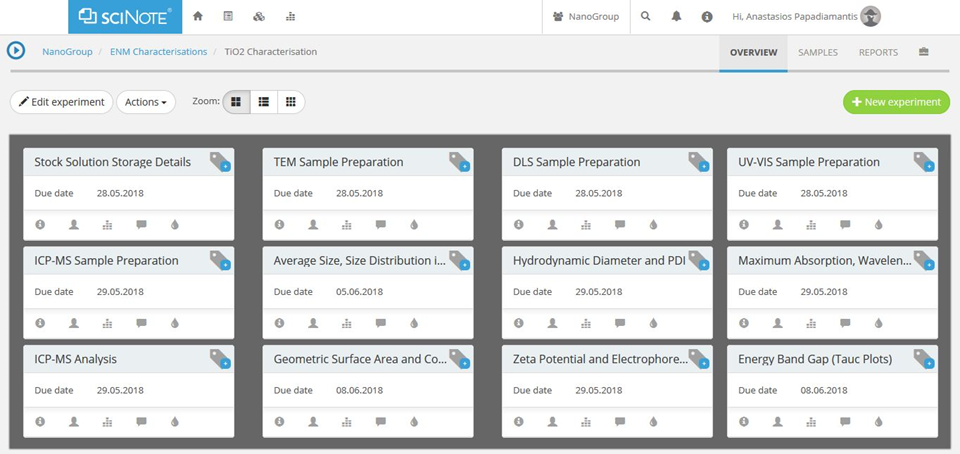
Figure A2. Experimental workflow for the physicochemical characterisation of an engineered nanomaterial (ENM) using the SciNote online lab-book.
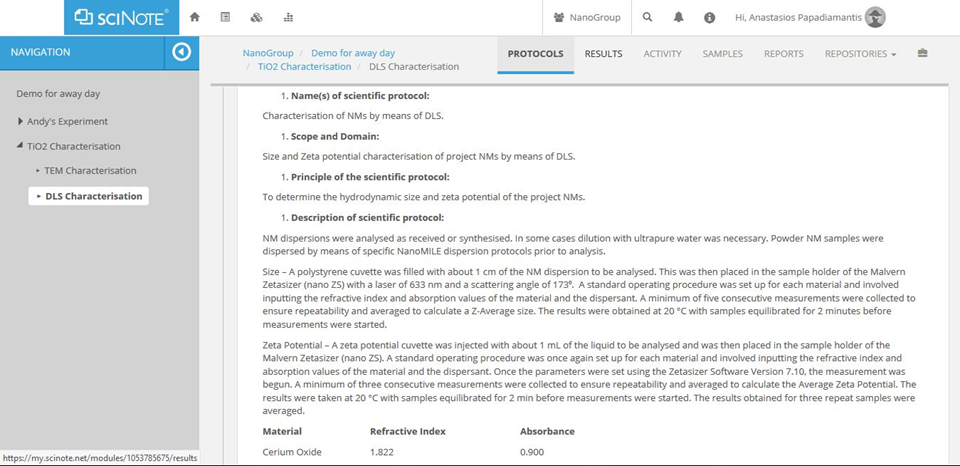
Figure A3. All experimental/computational workflow steps will be directly linked to detailed protocols.
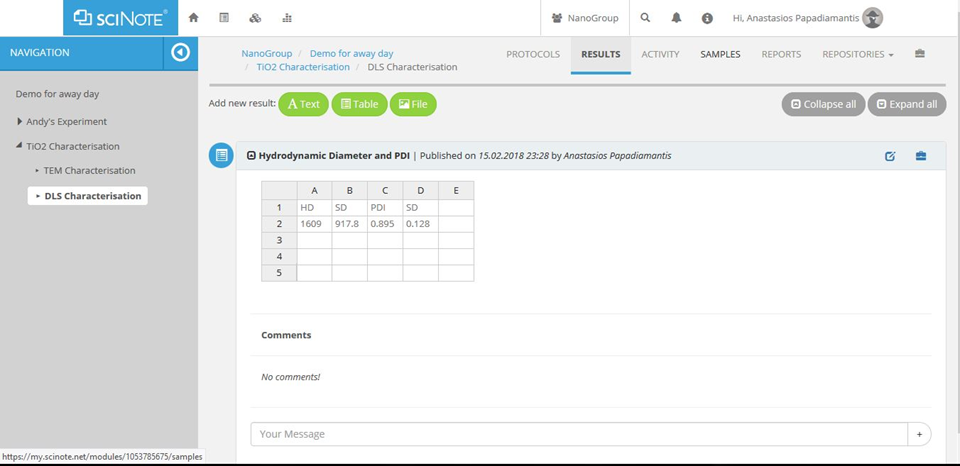
Figure A4. The results page will be where individual datasets acquired or calculated will be stored.
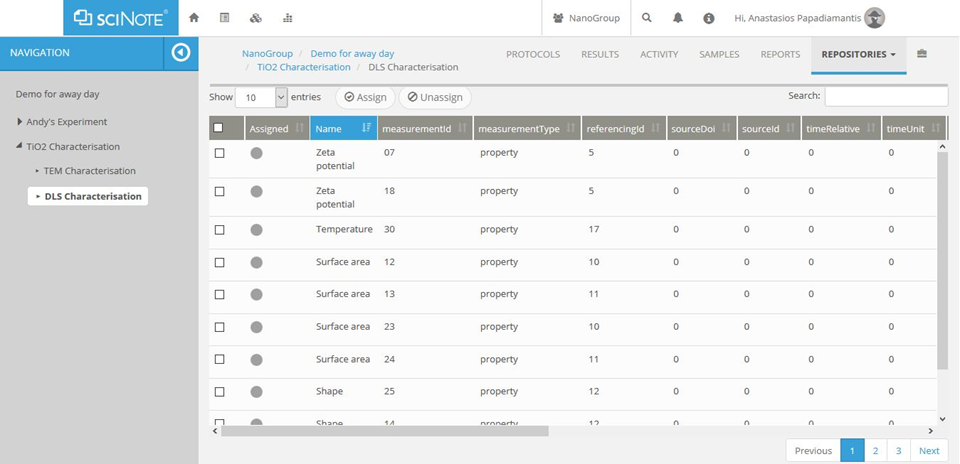
Figure A5. The complete set of acquired/computed data will be automatically
transferred to a curation template, which will be possible to automatically
extract and send for uploading to the
Step 2: Data acquisition and data curation
During the second step the actual data acquisition and simultaneous curation takes place along with data digitisation using either directly online lab-books described above or through the offline acquisition/analysis and subsequent transfer to the online lab-books and the specific data curation template.
Step 3: Data manipulation and data curation
The third step of the DMP will focus on data manipulation, i.e. the process
of data cleansing and control of the dataset’s quality and completeness.
During this stage the dataset completeness will be evaluated, potential
gaps and/or outliers identified and any repetition or supplemental data
acquisition needed will take place. When the dataset is completed it will
be stored into a data repository, compatible with EUON, and any tools
needed for the subsequent analysis will be implemented and linked to it.
The tools developed and on offer from the
Step 4: Data analysis
This step consists of all potential analytical or computational work
that needs to take place to the produced dataset, in order to extract
meaningful data interpretation and research outputs. The analysis will
be based on the needed tools provided via the
Step 5: Data Storage
Following the completion of the analytical or computational procedure, the produced data will be backed up and stored and the necessary metadata will be created. The metadata will be used to inform potential future users regarding the ownership of the datasets and user rights, information on retrieval and analytical descriptions of the respective datasets. During this step any necessary documentation describing the above will also be created and DOI numbers will be assigned to each dataset. DOIs will be used to assist with publications (submitting raw datasets to peer-reviewed journals in a digital form), referencing of the respective datasets by future users, to ensure that the original data owners are acknowledged and to facilitate the transition from closed to FAIR and Open data.
Step 6: FAIR Data
The final step in the
is agreement that data imported and generated as a result of JRA, NA and TA activities will be made Open and FAIR. For that, copyright and IP owners must be clearly defined.