workshops
Querying WikiPathways
Introduction
WikiPathways (https://www.wikipathways.org) is a biological pathway database known for its collaborative nature and open science approaches. The road toward a sustainable, community-driven pathway database goes through integration with other resources such as Wikidata and allowing more use, curation and redistribution of WikiPathways content. The SPARQL endpoint allows the access of the WikiPathways RDF and integrate its content with other databases. The RDF contains all pathways, their datanodes (genes, proteins, metabolites, etc.), author information, molecular descriptors, and more.
The WikiPathways SPARQL endpoint is accessible on https://sparql.wikipathways.org
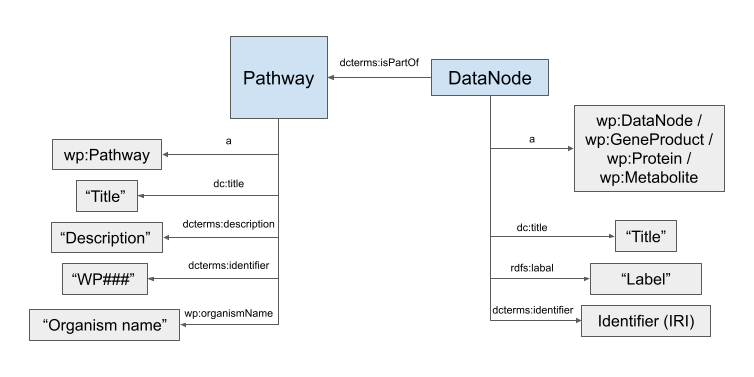
Figure of simplified RDF schema
Exercises
Exercise 1 - Listing of subjects
The simplest SPARQL queries to explore RDF is to retrieve full lists of subjects of a particular type, which is frequently defined with the predicate rdfs:type or a which can be used interchangably. See the below example of listing all pathways.
SELECT ?pathway
WHERE {
?pathway a wp:Pathway .
}
By looking at the RDF schema figure, you should be able to adapt the SPARQL query to answer all questions that follow.
- Question 1.1: What would we need to replace
wp:Pathwaywith to extract all datanodes? - wp:DataNode
- Question 1.2: What would we need to replace
wp:Pathwaywith to extract all metabolites? - wp:Metabolite
Because the WikiPathways RDF contains many properties of all subjects (such as pathways), we can also directly request all contents through the SPARQL query. For example, to extract the pathway title, we add ?pathway dc:title ?pathwaytitle to the SPARQL query and add ?pathwaytitle in the SELECT list. The returned table upon running the query will get wider, so you might need to scroll to the right to see it all.
- Question 1.3: What would we need to add to the query to extract the description of the pathway?
- Adding another variable to the
SELECTlist and requesting that variable by adding in the query?pathway dc:description ?[new variable name]. This should return a table with the added column. - Question 1.4: What would we need to add to the query to extract the identifiers of pathways?
- Adding another variable to the
SELECTlist and requesting that variable by adding in the query?pathway dcterms:identifier ?[new variable name]. This should return a table with the added column. - Question 1.5: What would we need to add to the query to extract to which organism name pathways apply to?
- Adding another variable to the
SELECTlist and requesting that variable by adding in the query?pathway wp:organismName ?[new variable name]. This should return a table with the added column.
Exercise 2 - Counting of subjects
This exercise is about creating simple SPARQL queries that count particular types of subjects in the RDF. See the example SPARQL query below that counts the number of pathways in the RDF.
SELECT (count (?pathway) as ?npathway)
WHERE {
?pathway a wp:Pathway .
}
When copying this SPARQL query and executing it, you will find that the WikiPathways contains 3094 pathways.
- Question 2.1: How many GeneProducts are present in the WikiPathways RDF?
- 37146
- Question 2.2: How many proteins are present in the WikiPathways RDF?
- 16145
Exercise 3 - More detailed exploration
With this exercise, the RDF will be explored a little more extensively. By combining statements in the RDF query, we can link multiple subjects and filter for content that we want to get back from the service. For example, the next query returns the title for pathway with ID WP1560:
SELECT ?pathwaytitle WHERE{
?pathway a wp:Pathway .
?pathway dc:title ?pathwaytitle .
?pathway dcterms:identifier "WP4868" .
}
- Question 3.1: What is the title of pathway with identifier WP5087?
- Malignant pleural mesothelioma
- Question 3.2: How many human (Homo sapiens) pathways are present in the WikiPathways RDF?
- 1224
- Question 3.3: Can you return the count of GeneProducts in pathway with ID WP78?
- 18
- Question 3.4: Which pathways have a gene with the label “BAP1”?
- WP4018 and WP5087
Challenge: construct a query that provides the count of DataNodes for each individual human pathway
- Link to SPARQL query: https://bit.ly/3lMqR3d
Exercise 4 - Federated SPARQL query
This final exercise adds an extra level of difficulty by linking the AOP-Wiki RDF with another database through SPARQL (this is called a Federated SPARQL query). In this exercise we will explore the connection between WikiPathways and AOP-Wiki. To do this exercise, you might want to do the AOP-Wiki SPARQL endpoint tutorial first.
The SPARQL query will need to contain a SERVICE function and the final query will have the following structure:
PREFIX aopo: <http://vocabularies.wikipathways.org/wp#>
SELECT [variables]
WHERE {
[query WikiPathways]
SERVICE <https://aopwiki.rdf.bigcat-bioinformatics.org/sparql> {
[query AOP-Wiki]
}}
- Question 4: What are the titles of Adverse Outcome Pathways in AOP-Wiki that are activated by metabolites with ChEBI IDs which are present in the pathway with identifier
WP5083in WikiPathways? In WikiPathways, you can extract the ChEBI ID using the predicatewp:bdbChEBIforwp:Metabolitesubjects. - Link to SPARQL query: https://bit.ly/3DBRMVN
End
Thank you for your participation. For any feedback or questions about this section, please contact Marvin Martens (marvin.martens@maastrichtuniversity.nl).