workshops
Querying the Nanosafety RDF
Introduction
Adverse Outcome Pathways (AOPs) have been proposed to explore interactions of chemicals/materials with biological systems. Each AOP starts with a molecular initiating event (MIE) and possibly ends with adverse outcome(s). So far, we do not fully understand how many nanomaterials interact with proteins, biomembranes, cells, and biological structures in general. There is limited insight into the toxicology-related key events (KEs) they trigger, such as oxidative stress and inflammation, or the biological processes underlying these KEs. For this SPARQL endpoint we integrate the annotation of nanomaterials and their MIEs with ontology annotation to demonstrate how we can then query AOPs and biological pathway information for these materials.
The SPARQL endpoint is available here: https://nanosafety.rdf.bigcat-bioinformatics.org/
Figure of RDF schema
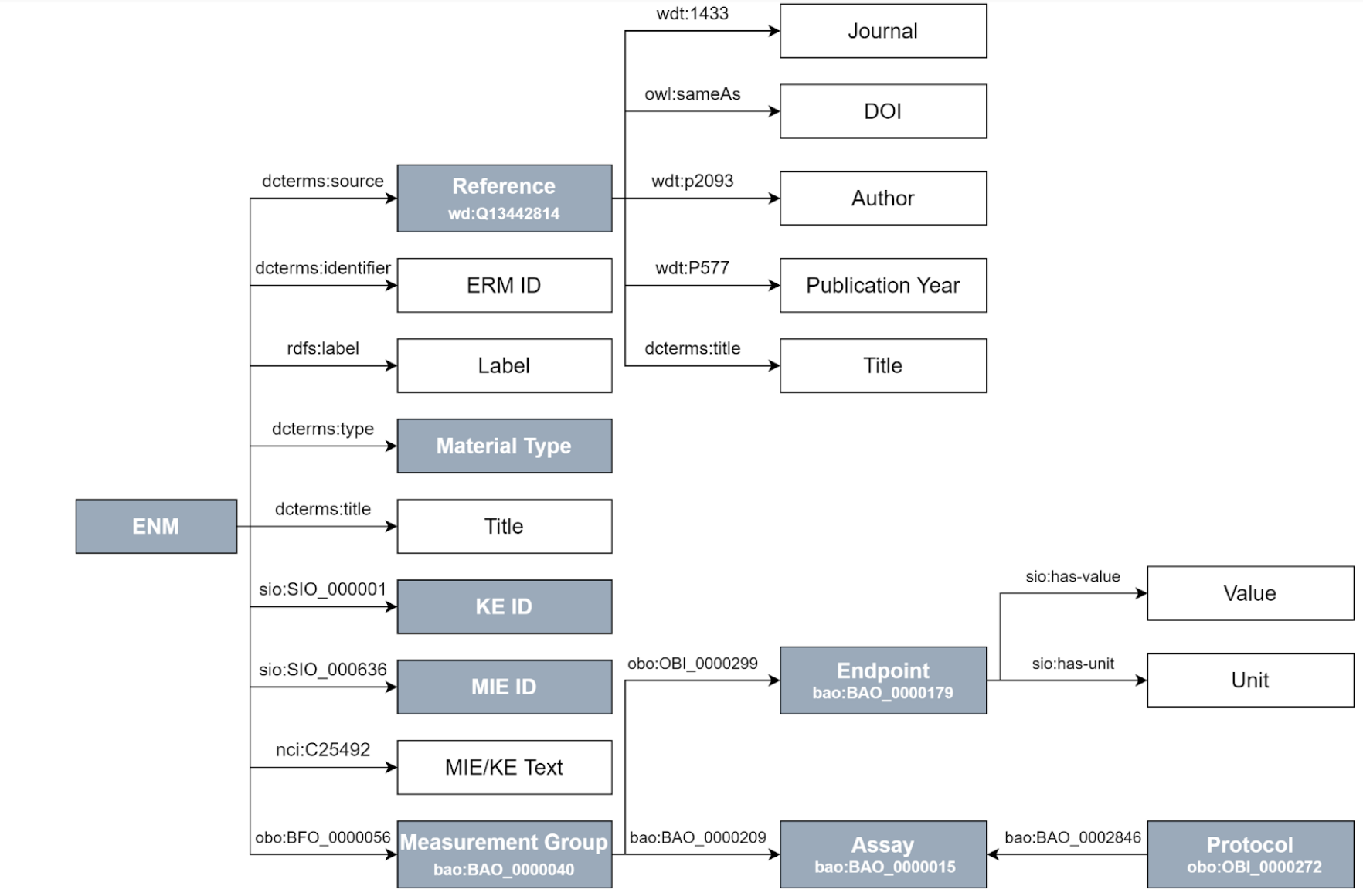
Graphical representation of the Resource Description Framework Schema for nanomaterials (NMs)
Exercises
Exercise 1 - listing of subjects
This first exercise is about creating simple SPARQL queries that list particular types of subjects in the RDF. See the example SPARQL query below to see the KE identifiers in the Nanosafety RDF.
PREFIX sio: <http://semanticscience.org/resource/>
select distinct ?KE where {
?s sio:SIO_000001 ?KE .
}
When copying this SPARQL query and executing it, you will see the KE id’s that are mentioned in the Nanosafety RDF. Of course these are hard to interpret. In the RDF schema you can see that the RDF also contains an MIE/KE text. This is the name of the KE as mentioned in the paper.
Modify the above query to select the MIE/KE text. Which predicate do you use? nci:C25492
Exercise 2 - counting of subjects
To get an overview of what is in the RDF you can create a number of queries to count thing. For example, how many distinct ERM identifiers are mentioned in the RDF? 87
And how many publications? 21
Can you count the number of endpoints? 7
How to count distict instances? Here is a hint: use COUNT(DISTINCT ?id as ?nid)
Exercise 3 - going one step further
As you can see in the RDF schema above you can extract information from the RDF about which endpoints a paper reports. Write a SPARQL query to list the endpoints mentioned in each paper (distinct DOI). Do all papers report all endpoints? No, quite a few only report 6 endpoints.
Exercise 4 - extracting values
Can you write a query that gives you the material label, the DOI and the value for the endpoint “shape”@en? What is the shape of “CeO2 NP-A”? spherical
The following query show you information about the medium in which the zeta potential is measured.
PREFIX bao: <http://www.bioassayontology.org/bao#>
PREFIX obo: <http://purl.obolibrary.org/obo/>
PREFIX sio: <http://semanticscience.org/resource/>
PREFIX enm: <http://purl.enanomapper.net/>
PREFIX dcterms: <http://purl.org/dc/terms/>
PREFIX wd: <http://www.wikidata.org/prop/direct/>
PREFIX owl: <http://www.w3.org/2002/07/owl#>
SELECT DISTINCT ?mlabel ?doi ?measurement ?medium WHERE {
?s rdfs:label ?mlabel ;
obo:BFO_0000056 ?mg .
?s dcterms:source ?source .
?source owl:sameAs ?doi .
?mg obo:OBI_0000299 ?o .
?o a bao:BAO_0000179 ;
rdfs:label ?measurement ;
rdfs:label "zeta potential"@en .
OPTIONAL{?o enm:has-condition ?g .}
OPTIONAL{?g rdfs:label ?medlabel
; sio:has-value ?medium .}
#OPTIONAL{value_range}
#OPTIONAL{value}
#OPTIONAL{unit}
} ORDER BY ?mlabel, ?doi
Add the lines for value_range, value and unit, so that the query also provides you with the values of the zeta potential in the different media. HINT: the predicate for value_range is not shown in the figure and is obo:STATO_0000035.
Run the query, what is the first material and value for the zeta potential if you ORDER BY the material name and DOI? 6 TiO2 NP A (anatase/rutile) with a value of -8.92 +/- 0.75 mV
Exercise 5 - Federated query
In exercise 1 we looked at the KE id’s that are mentioned in the Nanosafety RDF. Using a federated query to AOP wiki we can get the labels for these key events.
What is the label for KE https://identifiers.org/aop.events/1194? Increase, DNA damage
Here is a hint: use SERVICE -URL-{?mie dc:title ?mietitle .}
Bonus question
For each KE, can you find out which AOP it belongs to and what the associated AO is, using AOP wiki?
Answers
Example queries to get to the answers can be found here