workshops
Querying ChEMBL RDF
Introduction
ChEMBL is a manually curated database of bioactive molecules with drug-like properties. It brings together chemical, bioactivity and genomic data to aid the translation of genomic information into effective new drugs. Built upon the ChEMBL database, an RDF representation of the ChEMBL database is produced by the European Molecular Biology Laboratory - European Bioinformatics Institute (EMBL-EBI) and provided for download. The ChEMBL RDF model uses a basic internal ontology, called the ChEMBL Core Ontology (CCO), to identify all of the primary entities (e.g., Documents, Assays, Substances, Targets) in the ChEMBL database. The department of Bioinformatics (BiGCaT) at Maastricht University took the initiative to host the RDF and expose it to the scientific community through a SPARQL endpoint where queries can be executed against the RDF to find answers to biological questions. The tool is available through https://chemblmirror.rdf.bigcat-bioinformatics.org/.
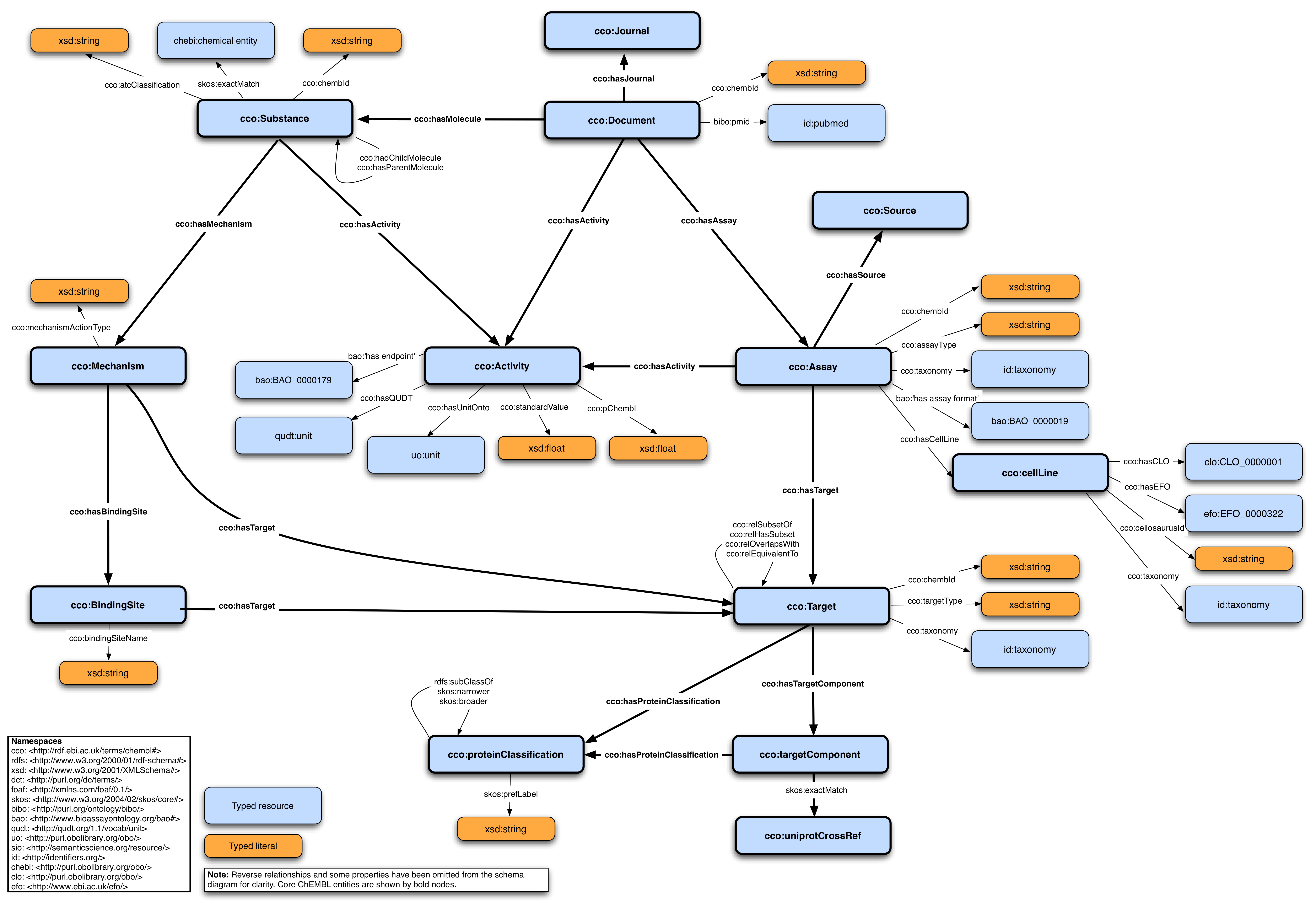
Figure of RDF schema
Exercises
Exercise 1 - Listing of subjects
The simplest SPARQL queries to explore RDF is to retrieve full lists of subjects of a particular type, which is frequently defined with the predicate rdfs:type or “a” which can be used interchangably. The type itself can be part of a hierarchy and then we can specify the type of a particular subclass using the predicate rdfs:subClassOf. See the below example of listing all molecules in the ChEMBL RDF where the molecule type is a subclass of the cco:Substance class.
Listing chemicals
PREFIX rdf: <http://www.w3.org/1999/02/22-rdf-syntax-ns#>
PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>
PREFIX owl: <http://www.w3.org/2002/07/owl#>
PREFIX xsd: <http://www.w3.org/2001/XMLSchema#>
PREFIX dc: <http://purl.org/dc/elements/1.1/>
PREFIX dcterms: <http://purl.org/dc/terms/>
PREFIX dbpedia2: <http://dbpedia.org/property/>
PREFIX dbpedia: <http://dbpedia.org/>
PREFIX foaf: <http://xmlns.com/foaf/0.1/>
PREFIX skos: <http://www.w3.org/2004/02/skos/core#>
PREFIX cco: <http://rdf.ebi.ac.uk/terms/chembl#>
SELECT ?molecule
WHERE {
?molecule a ?type .
?type rdfs:subClassOf* cco:Substance .
} limit 20
By looking at the RDF schema figure, you should be able to adapt the SPARQL query to answer the following question:
- Question 1.1: In order to get all targets in ChEMBL instead of substacnes, we need to replace the class
cco:Substancewith another one, which one is that? - cco:Target
Listing ChEMBL sources
PREFIX rdf: <http://www.w3.org/1999/02/22-rdf-syntax-ns#>
PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>
PREFIX owl: <http://www.w3.org/2002/07/owl#>
PREFIX xsd: <http://www.w3.org/2001/XMLSchema#>
PREFIX dc: <http://purl.org/dc/elements/1.1/>
PREFIX dcterms: <http://purl.org/dc/terms/>
PREFIX dbpedia2: <http://dbpedia.org/property/>
PREFIX dbpedia: <http://dbpedia.org/>
PREFIX foaf: <http://xmlns.com/foaf/0.1/>
PREFIX skos: <http://www.w3.org/2004/02/skos/core#>
PREFIX cco: <http://rdf.ebi.ac.uk/terms/chembl#>
SELECT ?Source
WHERE {
?Source ?p cco:Source .
} LIMIT 100
Take a look at the query above and answer the following question:
- Question 1.2: The previuos query gets all the sources in ChEMBL in a form of URLs, what do we need to add to the query to get the description of the source (using predicate: dcterms:description)?
- ?Source dcterms:description ?Description.
Exercise 2 - Counting of subjects
count assays that are used to get activity for the molecule CHEMBL294873
PREFIX rdf: <http://www.w3.org/1999/02/22-rdf-syntax-ns#>
PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>
PREFIX owl: <http://www.w3.org/2002/07/owl#>
PREFIX xsd: <http://www.w3.org/2001/XMLSchema#>
PREFIX dc: <http://purl.org/dc/elements/1.1/>
PREFIX dcterms: <http://purl.org/dc/terms/>
PREFIX dbpedia2: <http://dbpedia.org/property/>
PREFIX dbpedia: <http://dbpedia.org/>
PREFIX foaf: <http://xmlns.com/foaf/0.1/>
PREFIX skos: <http://www.w3.org/2004/02/skos/core#>
PREFIX cco: <http://rdf.ebi.ac.uk/terms/chembl#>
PREFIX chembl_molecule: <http://rdf.ebi.ac.uk/resource/chembl/molecule/>
SELECT (count(?assay) as ?assayCount)
WHERE {
?activity a cco:Activity ;
cco:hasMolecule chembl_molecule:CHEMBL294873 ;
cco:hasAssay ?assay .
?assay cco:hasSource ?source .
}
The query above gets the count of the assays used to measure the activity of the molecule with ID (CHEMBL294873). Knowing that, answer the following question:
- Question 2.1: How many assays you will get after running the query above?
- 6
- Question 2.2: What do you need to change in the previous query to get the count of the assays used to measure activity of molecule (CHEMBL46195) ?
- Change the molecule ID in the line “cco:hasMolecule chembl_molecule:CHEMBL294873 ;” from CHEMBL294873 to CHEMBL46195
- Question 2.3: Try the query after making the change, how many assays you get now?
- 2
Exercise 3 - More detailed exploration
Get all assay, binding affinity type (Kd, Ki, IC50) and affinity value for all compounds targeting Thrombin protein (CHEMBL204)
PREFIX chembl: <http://rdf.ebi.ac.uk/terms/chembl#>
PREFIX cco: <http://rdf.ebi.ac.uk/terms/chembl#>
PREFIX chembl_molecule: <http://rdf.ebi.ac.uk/resource/chembl/molecule/>
PREFIX chembl_target: <http://rdf.ebi.ac.uk/resource/chembl/target/>
SELECT distinct ?assayLabel ?assayType ?molLabel ?bindingAffinityType ?value WHERE {
?assay chembl:hasTarget chembl_target:CHEMBL204.
?activity chembl:hasAssay ?assay.
?assay cco:assayType ?assayType.
?activity chembl:hasMolecule ?molecule .
chembl_target:CHEMBL204 rdfs:label ?targetLabel.
?molecule rdfs:label ?molLabel.
?assay rdfs:label ?assayLabel.
VALUES ?bindingAffinityType {"Kd" "Ki" "IC50"}
?activity chembl:type ?bindingAffinityType.
?activity chembl:standardValue ?value.
} limit 100
The above query get assays and molecules information along with binding affinity type and value limited ti the top 100 entry. Knowing that, answer the following question:
- Question 3.1: How to limit the binding affinity type of the returned values to “IC50”?
- In the line “VALUES ?bindingAffinityType {“Kd” “Ki” “IC50”}”, remove “Ki” and “Kd” so only “IC50” is left between the curly brackets.
Exercise 4: assay, target and protein information
List all assays, target names and UniProt IDs for the drug Paracetamol (CHEBI:46195)
PREFIX rdf: <http://www.w3.org/1999/02/22-rdf-syntax-ns#>
PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>
PREFIX owl: <http://www.w3.org/2002/07/owl#>
PREFIX xsd: <http://www.w3.org/2001/XMLSchema#>
PREFIX dc: <http://purl.org/dc/elements/1.1/>
PREFIX dcterms: <http://purl.org/dc/terms/>
PREFIX dbpedia2: <http://dbpedia.org/property/>
PREFIX dbpedia: <http://dbpedia.org/>
PREFIX foaf: <http://xmlns.com/foaf/0.1/>
PREFIX skos: <http://www.w3.org/2004/02/skos/core#>
PREFIX cco: <http://rdf.ebi.ac.uk/terms/chembl#>
PREFIX chembl_molecule: <http://rdf.ebi.ac.uk/resource/chembl/molecule/>
SELECT ?activityLabel ?assayType ?targetLabel ?uniprot
WHERE {
?activity a cco:Activity ;
rdfs:label ?activityLabel;
cco:hasMolecule chembl_molecule:CHEMBL46195 ;
cco:hasAssay ?assay .
?assay cco:assayType ?assayType.
?assay cco:hasTarget ?target .
?target rdfs:label ?targetLabel.
?target cco:hasTargetComponent ?targetcmpt .
?targetcmpt cco:targetCmptXref ?uniprot .
?uniprot a cco:UniprotRef
}
The above query will get assay types, target and UniProt identifier for all the proteins tested fo binding with a single molecule (chembl_molecule:CHEMBL46195).
Modify the above query with a molecule of interest to you or one of the following molecules to see which proteins and what type of assays have been used with it.
- CHEMBL2094079
- CHEMBL334894
- CHEMBL62704
- CHEMBL553740
- CHEMBL108214
- CHEMBL33384
End
Thank you for your participation. For any feedback or questions about this section, please contact Ammar Ammar (a.ammar@maastrichtuniversity.nl).