workshops
Exercise 5: creating an eNanoMapper data set in RDF
| prev | toc |
In this final exercise, you are going to write a short data file capturing some minimal metadata about a recently published study around nanosafety.
Take the situation you published a new nanosafety study about some nanoparticle. And you want people to discover your new work if they search for studies on that nanoparticle. This Exercise will explain how to make your article more findable, fulfilling one of the requirements of FAIR data.
We basically start with a nanoparticle, the DOI of the article, and some info of who you are. This information can be submitted in various ways, but we here focus on a simple RDF-based file, which you can edit with a simple text editor, like Notepad. This write up is a shorter version of this longer tutorial.
- Step 1: Open a text editor (e.g.
Notepad.exeor TextMate)
After that, and because the document will be a valid RDF document, we need to specify a few shortcuts, which
are called namespaces in RDF documents.
- Step 2: Add the following namespace definitions to your document (or copy/paste this full template):
@prefix bao: <http://www.bioassayontology.org/bao#> .
@prefix cito: <http://purl.org/net/cito/> .
@prefix dc: <http://purl.org/dc/elements/1.1/> .
@prefix dcterms: <http://purl.org/dc/terms/> .
@prefix foaf: <http://xmlns.com/foaf/0.1/> .
@prefix npo: <http://purl.bioontology.org/ontology/npo#> .
@prefix obo: <http://purl.obolibrary.org/obo/> .
@prefix owner: <https://openrisknet.github.io/workshops/demo/owner/> .
@prefix rdf: <http://www.w3.org/1999/02/22-rdf-syntax-ns#> .
@prefix rdfs: <http://www.w3.org/2000/01/rdf-schema#> .
@prefix sio: <http://semanticscience.org/resource/> .
@prefix void: <http://rdfs.org/ns/void#> .
@prefix xsd: <http://www.w3.org/2001/XMLSchema#> .
Info about us
The first bit of information that a FAIR data set needs, is a bit of information about the data set.
- Step 3: Use the below template to describe your data set:
owner:DataSet1
a void:Dataset ;
dcterms:license <https://creativecommons.org/publicdomain/zero/1.0/> ;
dcterms:publisher "Egon Willighagen"@en ;
dcterms:description "Nanomaterials I am excited about."@en ;
dcterms:title "Exciting nanomaterials"@en .
- Step 4: Change the
dcterms:publisher,dcterms:description, anddcterms:titlevalues. For convenience, you can extend the description to include the citation to your research article.
Info about your nanomaterial
The next bit of information is about the nanomaterial. Here, the template looks like this:
owner:Substance1
a obo:CHEBI_59999 ;
rdfs:label "zinc oxide" ;
dcterms:source owner:DataSet1 ;
dcterms:type npo:NPO_1542 .
In this template, the obo:CHEBI_59999 is used to indicate that this resource is a chemical substance.
You can update the rdfs:label to match your material, and the dcterms:source says that this material
is part of the data set you created before.
Besides the label for the material, the second important thing to change here is the type of your
material. This is where the earlier exercise come in. There you looked up an eNanoMapper ontology IRI
or even minted a new IRI. This IRI is what you need for the dcterms:type line.
- Step 5: Update the material template for the name of your material and the IRI of your material
If you material has coatings, please refer to the longer tutorial for examples on how to do that.
Adding the size of your material
In the RDF discussed here, the data model of the BioAssay Ontology is reused for physicochemical
properties too. This model is very flexible and rich, but therefore also somewhat longer. We
define an assay (owner:particleSizeAssay), a measurement group (owner:Substance1_sizemg),
and finally the size (owner:Substance1_size):
owner:Substance1
obo:BFO_0000056 owner:Substance1_sizemg .
owner:particleSizeAssay
a npo:NPO_1694 , bao:BAO_0000015 ;
dc:title "Particle Size" ;
bao:BAO_0000209 owner:Substance1_sizemg .
owner:Substance1_sizemg a obo:BAO_0000040 ;
obo:OBI_0000299 owner:Substance1_size .
owner:Substance1_size a bao:BAO_0002128 ;
rdfs:label "primary particle size" ;
sio:has-unit "nm" ;
sio:has-value "13.6" .
In this template, basiclly only the resource IRIs ((owner:Substance1_sizemg) and (owner:Substance1_size))
change when you add more than one material.
But if you only want to describe during this workshop one nanomaterial, then you only need to change the actual size value, on the last line of this bit of template.
Step 6: Copy/paste this template code into your text editor, and update the size
Linking the particle to your article
The final step in creating the RDF is to link your particle description to the article where you studied your particle. This is the template, and note that the article is linked to the physicochemical data, rather than the particle:
owner:Substance1_size cito:usesDataFrom <https://doi.org/10.1021/es900754q> .
Step 7: Copy/paste this template and update the DOI in the IRI.
Uploading the RDF to the OpenRiskNet cloud
When done, explain your solution with your neighbor (peer review) and see if they agree with your formal description. You can also use this validation service to check the content of your file for syntax errors. The feedback from this service is easier to understand, than the below OpenRiskNet service will return.
When the file is done, you can then proceed to the following and upload your RDF into the OpenRiskNet platform:
- Step 8: Visit the OpenRiskNet eNanoMapper instance at http://nanomaterialdb-test.prod.openrisknet.org/ambit/
- Step 9: At the top, hover over
Data uploadand clickSpreadsheet upload. You should get a page that looks like this:
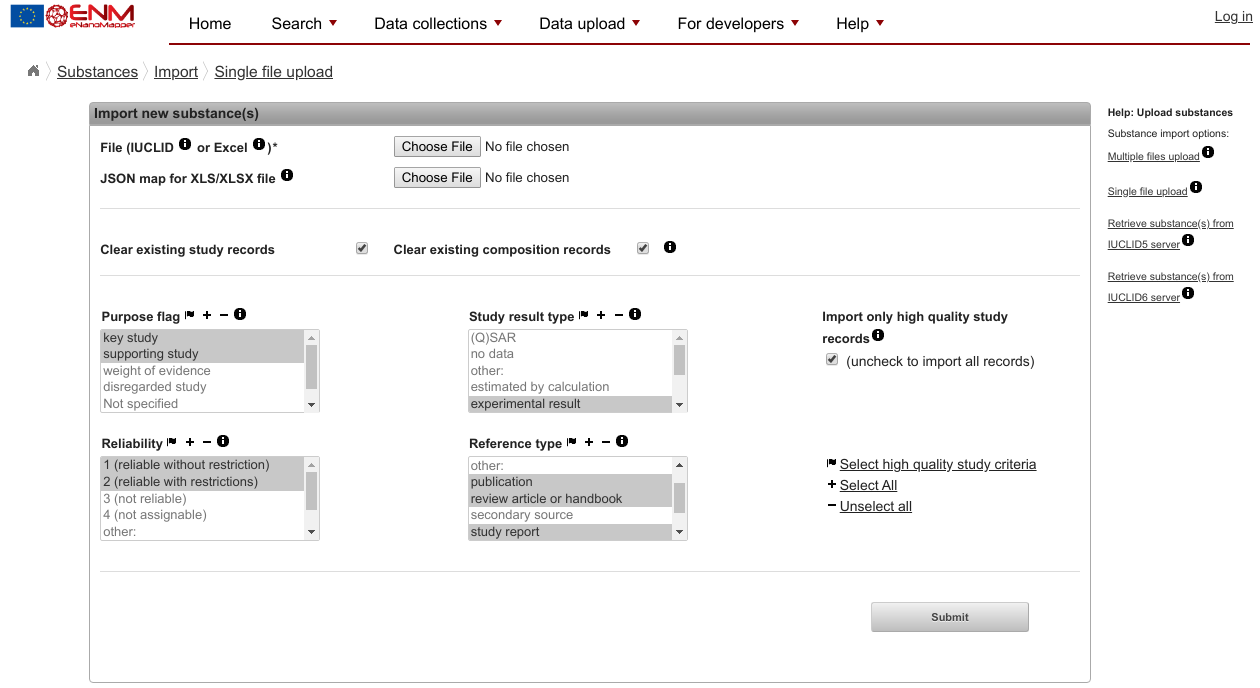
- Step 10: Ignore the mention of spreadsheets (it will load valid Turtle fine) and ignore all parameters on this page.
- Step 11: Use the top
Choose filebutton to select the file you created with your text editor - Step 12: Click Submit at the bottom
If all goes well, you should get a “Ready” message that looks like this:
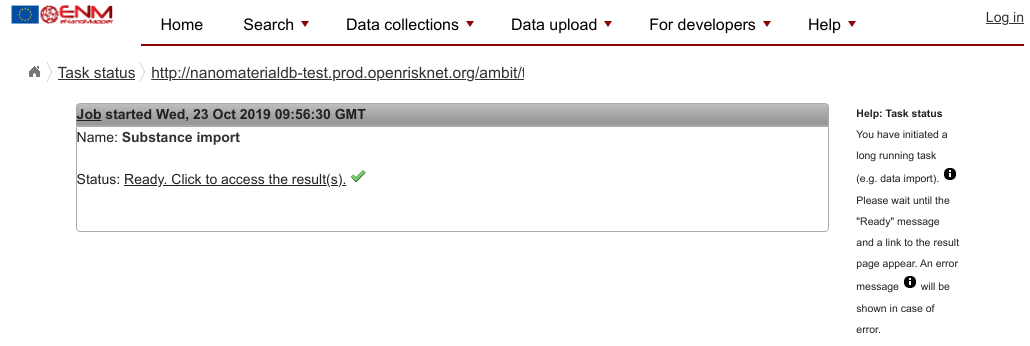
If you did not get a “Ready” message, then there is likely something wrong with your data file. Please discuss this with one of the Workshop assistents.
- Step 13: Visit http://nanomaterialdb-test.prod.openrisknet.org/ambit/substanceowner to confirm you see your data set
| prev | toc |
Copyright 2019 (C) Egon Willighagen - CC-BY Int. 4.0